IL-31 Uncouples Skin Inflammation from Itch Sensation in Allergic Dermatitis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Oncostatin M Exhibit Elevated Responsiveness to IL-31 Receptor
IL-31 Receptor (IL-31RA) Knockout Mice Exhibit Elevated Responsiveness to Oncostatin M This information is current as Janine Bilsborough, Sherri Mudri, Eric Chadwick, Brandon of September 28, 2021. Harder and Stacey R. Dillon J Immunol 2010; 185:6023-6030; Prepublished online 18 October 2010; doi: 10.4049/jimmunol.0902769 http://www.jimmunol.org/content/185/10/6023 Downloaded from References This article cites 29 articles, 6 of which you can access for free at: http://www.jimmunol.org/content/185/10/6023.full#ref-list-1 http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication by guest on September 28, 2021 *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2010 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology IL-31 Receptor (IL-31RA) Knockout Mice Exhibit Elevated Responsiveness to Oncostatin M Janine Bilsborough,1 Sherri Mudri,1 Eric Chadwick,2 Brandon Harder,3 and Stacey R. Dillon IL-31 signals through the heterodimeric receptor IL-31RA and oncostatin M receptor (OSMR), and has been linked with the development of atopic dermatitis, a Th2 cytokine-associated disease in humans. -

System, Method and Software for Calculation of a Cannabis Drug Efficiency Index for the Reduction of Inflammation
International Journal of Molecular Sciences Article System, Method and Software for Calculation of a Cannabis Drug Efficiency Index for the Reduction of Inflammation Nicolas Borisov 1,† , Yaroslav Ilnytskyy 2,3,†, Boseon Byeon 2,3,4,†, Olga Kovalchuk 2,3 and Igor Kovalchuk 2,3,* 1 Moscow Institute of Physics and Technology, 9 Institutsky lane, Dolgoprudny, Moscow Region 141701, Russia; [email protected] 2 Department of Biological Sciences, University of Lethbridge, Lethbridge, AB T1K 3M4, Canada; [email protected] (Y.I.); [email protected] (B.B.); [email protected] (O.K.) 3 Pathway Rx., 16 Sandstone Rd. S., Lethbridge, AB T1K 7X8, Canada 4 Biomedical and Health Informatics, Computer Science Department, State University of New York, 2 S Clinton St, Syracuse, NY 13202, USA * Correspondence: [email protected] † First three authors contributed equally to this research. Abstract: There are many varieties of Cannabis sativa that differ from each other by composition of cannabinoids, terpenes and other molecules. The medicinal properties of these cultivars are often very different, with some being more efficient than others. This report describes the development of a method and software for the analysis of the efficiency of various cannabis extracts to detect the anti-inflammatory properties of the various cannabis extracts. The method uses high-throughput gene expression profiling data but can potentially use other omics data as well. According to the signaling pathway topology, the gene expression profiles are convoluted into the signaling pathway activities using a signaling pathway impact analysis (SPIA) method. The method was tested by inducing inflammation in human 3D epithelial tissues, including intestine, oral and skin, and then exposing these tissues to various extracts and then performing transcriptome analysis. -

Supplementary Material DNA Methylation in Inflammatory Pathways Modifies the Association Between BMI and Adult-Onset Non- Atopic
Supplementary Material DNA Methylation in Inflammatory Pathways Modifies the Association between BMI and Adult-Onset Non- Atopic Asthma Ayoung Jeong 1,2, Medea Imboden 1,2, Akram Ghantous 3, Alexei Novoloaca 3, Anne-Elie Carsin 4,5,6, Manolis Kogevinas 4,5,6, Christian Schindler 1,2, Gianfranco Lovison 7, Zdenko Herceg 3, Cyrille Cuenin 3, Roel Vermeulen 8, Deborah Jarvis 9, André F. S. Amaral 9, Florian Kronenberg 10, Paolo Vineis 11,12 and Nicole Probst-Hensch 1,2,* 1 Swiss Tropical and Public Health Institute, 4051 Basel, Switzerland; [email protected] (A.J.); [email protected] (M.I.); [email protected] (C.S.) 2 Department of Public Health, University of Basel, 4001 Basel, Switzerland 3 International Agency for Research on Cancer, 69372 Lyon, France; [email protected] (A.G.); [email protected] (A.N.); [email protected] (Z.H.); [email protected] (C.C.) 4 ISGlobal, Barcelona Institute for Global Health, 08003 Barcelona, Spain; [email protected] (A.-E.C.); [email protected] (M.K.) 5 Universitat Pompeu Fabra (UPF), 08002 Barcelona, Spain 6 CIBER Epidemiología y Salud Pública (CIBERESP), 08005 Barcelona, Spain 7 Department of Economics, Business and Statistics, University of Palermo, 90128 Palermo, Italy; [email protected] 8 Environmental Epidemiology Division, Utrecht University, Institute for Risk Assessment Sciences, 3584CM Utrecht, Netherlands; [email protected] 9 Population Health and Occupational Disease, National Heart and Lung Institute, Imperial College, SW3 6LR London, UK; [email protected] (D.J.); [email protected] (A.F.S.A.) 10 Division of Genetic Epidemiology, Medical University of Innsbruck, 6020 Innsbruck, Austria; [email protected] 11 MRC-PHE Centre for Environment and Health, School of Public Health, Imperial College London, W2 1PG London, UK; [email protected] 12 Italian Institute for Genomic Medicine (IIGM), 10126 Turin, Italy * Correspondence: [email protected]; Tel.: +41-61-284-8378 Int. -

Knock-Out Validated Antibodies from Cloud-Clone Cat.No
Knock-out validated antibodies from Cloud-Clone Cat.No. Target PAA778Hu01 B-Cell Leukemia/Lymphoma 2 (Bcl2) PAL763Hu01 Myxovirus Resistance 1 (MX1) PAB698Hu01 Lactate Dehydrogenase B (LDHB) PAA009Hu01 Angiopoietin 2 (ANGPT2) PAA849Ra01 Glycogen Phosphorylase, Liver (PYGL) PAA153Hu01 Alpha-Fetoprotein (aFP) PAF460Hu01 Folate Receptor 1, Adult (FOLR1) PAB233Hu01 Cyclin Dependent Kinase 4 (CDK4) PAA150Hu04 Carcinoembryonic Antigen (CEA) PAB905Hu01 Interleukin 7 Receptor (IL7R) PAC823Hu01 Thymidine Kinase 1, Soluble (TK1) PAH838Hu01 Isocitrate Dehydrogenase 2, mitochondrial (IDH2) PAK078Mu01 Fas Associating Death Domain Containing Protein (FADD) PAA537Hu01 Enolase, Neuron Specific (NSE) PAA651Hu01 Hyaluronan Binding Protein 1 (HABP1) PAB215Hu02 Fibrinogen Beta (FGb) PAB769Hu01 S100 Calcium Binding Protein A6 (S100A6) PAB231Hu01 Keratin 18 (KRT18) PAH839Hu01 Isocitrate Dehydrogenase 1, Soluble (IDH1) PAE748Hu01 Karyopherin Alpha 2 (KPNa2) PAB081Hu02 Heat Shock 70kDa Protein 1A (HSPA1A) PAA778Mu01 B-Cell Leukemia/Lymphoma 2 (Bcl2) PAA853Hu03 Caspase 8 (CASP8) PAA399Mu01 High Mobility Group Protein 1 (HMG1) PAA303Mu01 Galectin 3 (GAL3) PAA537Mu02 Enolase, Neuron Specific (NSE) PAA994Ra01 Acid Phosphatase 1 (ACP1) PAB083Ra01 Superoxide Dismutase 2, Mitochondrial (SOD2) PAB449Mu01 Enolase, Non Neuronal (NNE) PAA376Mu01 Actinin Alpha 2 (ACTN2) PAA553Ra01 Matrix Metalloproteinase 9 (MMP9) PAA929Bo01 Retinol Binding Protein 4, Plasma (RBP4) PAA491Ra02 Keratin 2 (KRT2) PAC025Hu01 Keratin 8 (KRT8) PAB231Mu01 Keratin 18 (KRT18) PAC598Hu03 Vanin 1 (VNN1) -

Evolutionary Divergence and Functions of the Human Interleukin (IL) Gene Family Chad Brocker,1 David Thompson,2 Akiko Matsumoto,1 Daniel W
UPDATE ON GENE COMPLETIONS AND ANNOTATIONS Evolutionary divergence and functions of the human interleukin (IL) gene family Chad Brocker,1 David Thompson,2 Akiko Matsumoto,1 Daniel W. Nebert3* and Vasilis Vasiliou1 1Molecular Toxicology and Environmental Health Sciences Program, Department of Pharmaceutical Sciences, University of Colorado Denver, Aurora, CO 80045, USA 2Department of Clinical Pharmacy, University of Colorado Denver, Aurora, CO 80045, USA 3Department of Environmental Health and Center for Environmental Genetics (CEG), University of Cincinnati Medical Center, Cincinnati, OH 45267–0056, USA *Correspondence to: Tel: þ1 513 821 4664; Fax: þ1 513 558 0925; E-mail: [email protected]; [email protected] Date received (in revised form): 22nd September 2010 Abstract Cytokines play a very important role in nearly all aspects of inflammation and immunity. The term ‘interleukin’ (IL) has been used to describe a group of cytokines with complex immunomodulatory functions — including cell proliferation, maturation, migration and adhesion. These cytokines also play an important role in immune cell differentiation and activation. Determining the exact function of a particular cytokine is complicated by the influence of the producing cell type, the responding cell type and the phase of the immune response. ILs can also have pro- and anti-inflammatory effects, further complicating their characterisation. These molecules are under constant pressure to evolve due to continual competition between the host’s immune system and infecting organisms; as such, ILs have undergone significant evolution. This has resulted in little amino acid conservation between orthologous proteins, which further complicates the gene family organisation. Within the literature there are a number of overlapping nomenclature and classification systems derived from biological function, receptor-binding properties and originating cell type. -

Differentially Methylated Genes
10/30/2013 Disclosures Key Rheumatoid Arthritis-Associated Pathogenic Pathways Revealed by Integrative Analysis of RA Omics Datasets Consultant: IGNYTA Funding: Rheumatology Research Foundation By John W. Whitaker, Wei Wang and Gary S. Firestein DNA methylation and gene regulation The RA methylation signature in FLS DNA methylation – DNMT1 (maintaining methylation) OA – DNMT3a, 3b (de novo methylation) RA % of CpG methylation: 0% 100% Nakano et al. 2013 ARD AA06 AANAT AARS ABCA6 ABCC12 ABCG1 ABHD8 ABL2 ABR ABRA ACACA ACAN ACAP3 ACCSL ACN9 ACOT7 ACOX2 ACP5 ACP6 ACPP ACSL1 ACSL3 ACSM5 ACVRL1 ADAM10 ADAM32 ADAM33 ADAMTS12 ADAMTS15 ADAMTS19 ADAMTS4 ADAT3 ADCK4 ADCK5 ADCY2 ADCY3 ADCY6 ADORA1 ADPGK ADPRHL1 ADTRP AFAP1 AFAP1L2 AFF3 AFG3L1P AGAP11 AGER AGTR1 AGXT AIF1L AIM2 AIRE AJUBA AK4 AKAP12 AKAP2 AKR1C2 AKR1E2 AKT2 ALAS1 ALDH1L1-AS1 ALDH3A1 ALDH3B1 ALDH8A1 ALDOB ALDOC ALOX12 ALPK3 ALS2CL ALX4 AMBRA1 AMPD2 AMPD3 ANGPT1 ANGPT2 ANGPTL5 ANGPTL6 ANK1 ANKMY2 ANKRD29 ANKRD37 ANKRD53 ANO3 ANO6 ANO7 ANP32C ANXA6 ANXA8L2 AP1G1 AP2A2 AP2M1 AP5B1 APBA2 APC APCDD1 APOBEC3B APOBEC3G APOC1 APOH APOL6 APOLD1 APOM AQP1 AQP10 AQP6 AQP9 ARAP1 ARHGAP24 ARHGAP42 ARHGEF19 ARHGEF25 ARHGEF3 ARHGEF37 ARHGEF7 ARL4C ARL6IP 5 ARL8B ARMC3 ARNTL2 ARPP21 ARRB1 ARSI ASAH2B ASB10 ASB2 ASCL2 ASIC4 ASPH ATF3 ATF7 ATL1 ATL3 ATP10A ATP1A1 ATP1A4 ATP2C1 ATP5A1 ATP5EP2 ATP5L2 ATP6V0CP3 ATP6V1C1 ATP6V1E2 ATXN7L1 ATXN7L2 AVPI1 AXIN2 B3GNT7 B3GNT8 B3GNTL1 BACH1 BAG3 Differential methylated genes in RA FLS BAIAP2L2 BANP BATF BATF2 BBS2 BCAS4 BCAT1 BCL7C BDKRB2 BEGAIN BEST1 BEST3 -

Human Colon Cancer Primer Library
Human Colon Cancer Primer Library Catalog No: HCCR-1 Supplier: RealTimePrimers Lot No: XXXXX Supplied as: solid Stability: store at -20°C Description Contains 88 primer sets directed against cytokine and chemokine receptor genes and 8 housekeeping gene primer sets. Provided in a 96-well microplate (20 ul - 10 uM). Perform up to 100 PCR arrays (based on 20 ul assay volume per reaction). Just add cDNA template and SYBR green master mix. Gene List: • CCR1 chemokine (C-C motif) receptor 1 • IL11RA interleukin 11 receptor, alpha • CCR2 chemokine (C-C motif) receptor 2 • IL11RB interleukin 11 receptor, beta • CCR3 chemokine (C-C motif) receptor 3 • IL12RB1 interleukin 12 receptor, beta 1 • CCR4 chemokine (C-C motif) receptor 4 • IL12RB2 interleukin 12 receptor, beta 2 • CCR5 chemokine (C-C motif) receptor 5 • IL13RA1 interleukin 13 receptor, alpha 1 • CCR6 chemokine (C-C motif) receptor 6 • IL13RA2 interleukin 13 receptor, alpha 2 • CCR7 chemokine (C-C motif) receptor 7 • IL15RA interleukin 15 receptor, alpha • CCR8 chemokine (C-C motif) receptor 8 • IL15RB interleukin 15 receptor, beta • CCR9 chemokine (C-C motif) receptor 9 • IL17RA interleukin 17 receptor A • CCR10 chemokine (C-C motif) receptor 10 • IL17RB interleukin 17 receptor B • CX3CR1 chemokine (C-X3-C motif) receptor 1 • IL17RC interleukin 17 receptor C • CXCR1 chemokine (C-X-C motif) receptor 1 • IL17RD interleukin 17 receptor D • CXCR2 chemokine (C-X-C motif) receptor 2 • IL17RE interleukin 17 receptor E • CXCR3 chemokine (C-X-C motif) receptor 3 • IL18R1 interleukin 18 receptor -

Epigenetic Mechanisms Are Involved in the Oncogenic Properties of ZNF518B in Colorectal Cancer
Epigenetic mechanisms are involved in the oncogenic properties of ZNF518B in colorectal cancer Francisco Gimeno-Valiente, Ángela L. Riffo-Campos, Luis Torres, Noelia Tarazona, Valentina Gambardella, Andrés Cervantes, Gerardo López-Rodas, Luis Franco and Josefa Castillo SUPPLEMENTARY METHODS 1. Selection of genomic sequences for ChIP analysis To select the sequences for ChIP analysis in the five putative target genes, namely, PADI3, ZDHHC2, RGS4, EFNA5 and KAT2B, the genomic region corresponding to the gene was downloaded from Ensembl. Then, zoom was applied to see in detail the promoter, enhancers and regulatory sequences. The details for HCT116 cells were then recovered and the target sequences for factor binding examined. Obviously, there are not data for ZNF518B, but special attention was paid to the target sequences of other zinc-finger containing factors. Finally, the regions that may putatively bind ZNF518B were selected and primers defining amplicons spanning such sequences were searched out. Supplementary Figure S3 gives the location of the amplicons used in each gene. 2. Obtaining the raw data and generating the BAM files for in silico analysis of the effects of EHMT2 and EZH2 silencing The data of siEZH2 (SRR6384524), siG9a (SRR6384526) and siNon-target (SRR6384521) in HCT116 cell line, were downloaded from SRA (Bioproject PRJNA422822, https://www.ncbi. nlm.nih.gov/bioproject/), using SRA-tolkit (https://ncbi.github.io/sra-tools/). All data correspond to RNAseq single end. doBasics = TRUE doAll = FALSE $ fastq-dump -I --split-files SRR6384524 Data quality was checked using the software fastqc (https://www.bioinformatics.babraham. ac.uk /projects/fastqc/). The first low quality removing nucleotides were removed using FASTX- Toolkit (http://hannonlab.cshl.edu/fastxtoolkit/). -

IL-31 Expression by Inflammatory Cells Is Preferentially Elevated In
Acta Derm Venereol 2012; 92: 24–28 INVESTIGATIVE REPORT IL-31 Expression by Inflammatory Cells is Preferentially Elevated in Atopic Dermatitis Stephan NOBBE1, Piotr DZIUNYCZ1, Beda MÜHLEISEN1, Janine BILSBOROUGH2, Stacey R. DILLON2, Lars E. FRENCH1 and Günther F. L. HOFBAUER1 1Department of Dermatology, University Hospital Zürich, Switzerland, and 2Department of Immunology, ZymoGenetics, Inc., WA, Seattle, USA Interleukin-31 (IL-31) is a recently discovered cytokine STAT and PI-3 kinase signaling pathways (1, 5, 6). The re- expressed in many human tissues, and predominantly by ceptor components IL-31RA and OSMR are co-expressed activated CD4+ T cells. IL-31 signals through a hetero- on keratinocytes and activated monocytes (1). Besides the dimeric receptor consisting of IL-31 receptor alpha (IL- skin, other tissues, such as testis, thymus, trachea, intestinal 31RA) and oncostatin M receptor beta (OSMR). Earlier epithelial cells and dorsal root ganglia, show mRNA co- studies have shown involvement of IL-31 and its receptor expression for the receptor components (1, 3, 7, 8). components IL-31RA and OSMR in atopic dermatitis, Atopic dermatitis (AD) is a chronic inflammatory skin pruritus and Th2-weighted inflammation at the mRNA disease that is associated with intense pruritus. The me- level. The aim of this study was to investigate IL-31 pro- chanisms underlying dermatitis and pruritus are complex tein expression in skin of such conditions. Immunohisto- and not fully understood (9, 10), but there is emerging chemical staining for IL-31, IL-31RA and OSMR was evidence that IL-31 may play a role. Transgenic mice performed in formalin-fixed paraffin-embedded biopsy that overexpress IL-31 develop severe pruritus and skin specimens. -

Radiation Response and Regulation of Apoptosis
Author's personal copy EXPERIMENTAL CELL RESEARCH 317 (2011) 1548– 1566 available at www.sciencedirect.com www.elsevier.com/locate/yexcr Research Article Radiation response and regulation of apoptosis induced by a combination of TRAIL and CHX in cells lacking mitochondrial DNA: A role for NF-κB–STAT3-directed gene expression Vladimir N. Ivanov⁎,1, Shanaz A. Ghandhi1, Hongning Zhou, Sarah X. Huang, Yunfei Chai, Sally A. Amundson, Tom K. Hei Center for Radiological Research, Department of Radiation Oncology, College of Physicians and Surgeons, Columbia University, New York, NY 10032, USA ARTICLE INFORMATION ABSTRACT Article Chronology: Mitochondrial DNA depleted (ρ0) human skin fibroblasts (HSF) with suppressed oxidative Received 8 December 2010 phosphorylation were characterized by significant changes in the expression of 2100 nuclear Revised version received genes, encoding numerous protein classes, in NF-κB and STAT3 signaling pathways, and by 16 February 2011 decreased activity of mitochondrial death pathway, compared to the parental ρ+ HSF. In contrast, Accepted 14 March 2011 the extrinsic TRAIL/TRAIL-Receptor mediated death pathway remained highly active, and Available online 2 April 2011 exogenous TRAIL in a combination with cycloheximide (CHX) induced higher levels of apoptosis in ρ0 cells compared to ρ+ HSF. Global gene expression analysis using microarray and Keywords: qRT-PCR demonstrated that mRNA expression levels of many growth factors and their adaptor Mitochondria proteins (FGF13, HGF, IGFBP4, IGFBP6, and IGFL2), cytokines (IL6, ΙL17Β, ΙL18, ΙL19, and ΙL28Β) and NF-κB cytokine receptors (IL1R1, IL21R, and IL31RA) were substantially decreased after mitochondrial STAT3 DNA depletion. Some of these genes were targets of NF-κB and STAT3, and their protein products IL6 could regulate the STAT3 signaling pathway. -
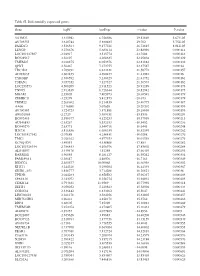
Table SI. Differentially Expressed Genes. Gene Logfc Aveexpr T Value
Table SI. Differentially expressed genes. Gene logFC AveExpr t value P‑value VSTM2L 3.315942 6.756386 29.82869 5.67E‑05 AV703555 3.610744 5.880485 29.702 5.75E‑05 FAXDC2 3.526564 9.177221 26.72815 8.01E‑05 LENG9 3.270676 5.683114 23.88996 0.000114 LOC101927507 ‑2.94917 3.515774 ‑23.7604 0.000116 BC028967 3.56197 4.208635 22.95804 0.000129 TMEM27 3.010875 8.053376 22.81882 0.000132 QPRT 2.56467 7.272755 22.37587 0.00014 TBC1D2 3.789191 6.823331 21.58772 0.000157 AF085835 2.602455 4.690477 21.43982 0.00016 TYROBP 2.386592 5.236329 21.41752 0.000161 TSPAN1 3.057352 7.127327 21.36574 0.000162 LOC253573 4.500209 2.911523 20.91289 0.000173 TNNC1 2.913839 6.726644 20.82942 0.000175 MGAM 2.35805 7.405973 20.68591 0.000179 ZDHHC11 ‑3.25279 5.412875 ‑20.673 0.000179 TRIM22 2.268602 8.214839 20.40775 0.000187 AA06 2.716698 5.07658 20.20202 0.000193 AV700385 3.724723 5.871881 20.18696 0.000193 AW851069 ‑2.2729 5.389451 ‑19.8536 0.000204 BC002465 2.590377 4.125253 19.57939 0.000213 AU146893 ‑2.63267 5.532153 ‑18.9452 0.000236 BC040270 ‑2.63771 5.540716 ‑18.6441 0.000248 H28731 3.816856 6.686159 18.32359 0.000262 LOC101927342 ‑2.07689 4.258431 ‑18.0208 0.000276 TMC1 2.028162 4.977353 18.01518 0.000276 KCNQ1DN ‑3.49551 4.538808 ‑17.883 0.000282 LOC101926934 2.786453 4.106976 17.85802 0.000284 AL043897 3.449478 4.51258 17.66439 0.000293 RARRES1 1.96055 8.281127 16.95242 0.000334 FAM189A1 ‑2.38147 4.48506 ‑16.7163 0.000349 IGDCC4 2.385577 10.00068 16.66958 0.000352 KLK11 3.244328 5.290104 16.44339 0.000367 GRID1‑AS1 1.858777 5.714288 16.26821 0.00038 UPK3B -

Final Korrekturen 3
The role of IL-31 in skin barrier formation Von der Fakultät für Mathematik, Informatik und Naturwissenschaften der RWTH Aachen University zur Erlangung des akademischen Grades eines Doktors der Naturwissenschaften genehmigte Dissertation vorgelegt von Master of Science Biotechnologie Kai Herbert Hänel aus Neuss Berichter: Universitätsprofessor Dr. rer. nat. Bernd Lüscher apl. Prof. Dr. rer. nat. Dirk Ostareck Tag der mündlichen Prüfung: 26.05.2015 Diese Dissertation ist auf den Internetseiten der Hochschulbibliothek online verfügbar. II No man should escape our universities without knowing how little he knows. J. Robert Oppenheimer Parts of this thesis were prepublished Hänel, K.H., Cornelissen, C., Lüscher, B., and Baron, J.M. (2013). Cytokines and the Skin Barrier. International Journal of Molecular Sciences, 14(4), 6720–6745. Hänel, K.H ., Cornelissen, C., Amann, P.M., Marquardt, Y., Czaja, K., Kim, A., Bickers, D.R., Lüscher, B. and Baron, J.M. Control of the mechanical and antimicrobial skin barrier by an IL-31 – IL-1 axis (manuscript submitted) III Abstract The skin is the largest organ in the human body and its major function is to protect from harmful environmental influences and to prevent dehydration. It forms the first barrier against biological, chemical and physical stress. Atopic dermatitis (AD) is a relapsing inflammatory skin disease. Hallmarks of AD are pruritic, eczematous erythematous plaques resulting in reduced skin barrier function, increasing transepidermal water loss and, affected by this, dry skin. AD commonly begins in early childhood and affects 10-20% of children and 1-3% of adults in industrialized countries. AD patients show significant higher levels of IL-31 in serum and increased IL31 mRNA levels in lesional skin samples.