Expression Profiling of Macrophages Reveals Multiple Populations With
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Screening and Identification of Key Biomarkers in Clear Cell Renal Cell Carcinoma Based on Bioinformatics Analysis
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Screening and identification of key biomarkers in clear cell renal cell carcinoma based on bioinformatics analysis Basavaraj Vastrad1, Chanabasayya Vastrad*2 , Iranna Kotturshetti 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. 3. Department of Ayurveda, Rajiv Gandhi Education Society`s Ayurvedic Medical College, Ron, Karnataka 562209, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Clear cell renal cell carcinoma (ccRCC) is one of the most common types of malignancy of the urinary system. The pathogenesis and effective diagnosis of ccRCC have become popular topics for research in the previous decade. In the current study, an integrated bioinformatics analysis was performed to identify core genes associated in ccRCC. An expression dataset (GSE105261) was downloaded from the Gene Expression Omnibus database, and included 26 ccRCC and 9 normal kideny samples. Assessment of the microarray dataset led to the recognition of differentially expressed genes (DEGs), which was subsequently used for pathway and gene ontology (GO) enrichment analysis. -

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

Single Cell Transcriptome Atlas of Immune Cells in Human Small
bioRxiv preprint doi: https://doi.org/10.1101/721258; this version posted August 1, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Single cell transcriptome atlas of immune cells in human small 2 intestine and in celiac disease 3 4 Nader Atlasy1,a,4, Anna Bujko2,4, Peter B Brazda1,a, Eva Janssen-Megens1,a , Espen S. 5 Bækkevold2, Jørgen Jahnsen3, Frode L. Jahnsen2, Hendrik G. Stunnenberg1,a,* 6 7 8 1Department of Molecular Biology, Science Faculty, Radboud University, Nijmegen, The 9 Netherlands. 10 2 Department of Pathology, University of Oslo and Oslo University Hospital, Rikshospitalet, 11 Oslo, Norway 12 3 Department of Gastroenterology, Akershus University Hospital and University of Oslo, 13 Oslo, Norway. 14 15 acurrent address: Princess Maxima Centre for Pediatric Oncology, Heidelberglaan 25, 3584 16 CS Utrecht 17 18 19 4These authors contributed equally to this study 20 21 22 *Corresponding author: [email protected] 23 24 25 26 Celiac disease (CeD) is an autoimmune disorder in which ingestion of dietary gluten 27 triggers an immune reaction in the small intestine1,2. The CeD lesion is characterized by 28 crypt hyperplasia, villous atrophy and chronic inflammation with accumulation of 29 leukocytes both in the lamina propria (LP) and in the epithelium3, which eventually 30 leads to destruction of the intestinal epithelium1 and subsequent digestive complications 31 and higher risk of non-hodgkin lymphoma4. A lifetime gluten-free diet is currently the 32 only available treatment5. Gluten-specific LP CD4 T cells and cytotoxic intraepithelial 33 CD8+ T cells are thought to be central in disease pathology1,6-8, however, CeD is a 34 complex immune-mediated disorder and to date the findings are mostly based on 35 analysis of heterogeneous cell populations and on animal models. -

List of Genes Used in Cell Type Enrichment Analysis
List of genes used in cell type enrichment analysis Metagene Cell type Immunity ADAM28 Activated B cell Adaptive CD180 Activated B cell Adaptive CD79B Activated B cell Adaptive BLK Activated B cell Adaptive CD19 Activated B cell Adaptive MS4A1 Activated B cell Adaptive TNFRSF17 Activated B cell Adaptive IGHM Activated B cell Adaptive GNG7 Activated B cell Adaptive MICAL3 Activated B cell Adaptive SPIB Activated B cell Adaptive HLA-DOB Activated B cell Adaptive IGKC Activated B cell Adaptive PNOC Activated B cell Adaptive FCRL2 Activated B cell Adaptive BACH2 Activated B cell Adaptive CR2 Activated B cell Adaptive TCL1A Activated B cell Adaptive AKNA Activated B cell Adaptive ARHGAP25 Activated B cell Adaptive CCL21 Activated B cell Adaptive CD27 Activated B cell Adaptive CD38 Activated B cell Adaptive CLEC17A Activated B cell Adaptive CLEC9A Activated B cell Adaptive CLECL1 Activated B cell Adaptive AIM2 Activated CD4 T cell Adaptive BIRC3 Activated CD4 T cell Adaptive BRIP1 Activated CD4 T cell Adaptive CCL20 Activated CD4 T cell Adaptive CCL4 Activated CD4 T cell Adaptive CCL5 Activated CD4 T cell Adaptive CCNB1 Activated CD4 T cell Adaptive CCR7 Activated CD4 T cell Adaptive DUSP2 Activated CD4 T cell Adaptive ESCO2 Activated CD4 T cell Adaptive ETS1 Activated CD4 T cell Adaptive EXO1 Activated CD4 T cell Adaptive EXOC6 Activated CD4 T cell Adaptive IARS Activated CD4 T cell Adaptive ITK Activated CD4 T cell Adaptive KIF11 Activated CD4 T cell Adaptive KNTC1 Activated CD4 T cell Adaptive NUF2 Activated CD4 T cell Adaptive PRC1 Activated -

Human ADAM12 Quantikine ELISA
Quantikine® ELISA Human ADAM12 Immunoassay Catalog Number DAD120 For the quantitative determination of A Disintegrin And Metalloproteinase domain- containing protein 12 (ADAM12) concentrations in cell culture supernates, serum, plasma, and urine. This package insert must be read in its entirety before using this product. For research use only. Not for use in diagnostic procedures. TABLE OF CONTENTS SECTION PAGE INTRODUCTION .....................................................................................................................................................................1 PRINCIPLE OF THE ASSAY ...................................................................................................................................................2 LIMITATIONS OF THE PROCEDURE .................................................................................................................................2 TECHNICAL HINTS .................................................................................................................................................................2 MATERIALS PROVIDED & STORAGE CONDITIONS ...................................................................................................3 OTHER SUPPLIES REQUIRED .............................................................................................................................................3 PRECAUTIONS .........................................................................................................................................................................4 -

Seq2pathway Vignette
seq2pathway Vignette Bin Wang, Xinan Holly Yang, Arjun Kinstlick May 19, 2021 Contents 1 Abstract 1 2 Package Installation 2 3 runseq2pathway 2 4 Two main functions 3 4.1 seq2gene . .3 4.1.1 seq2gene flowchart . .3 4.1.2 runseq2gene inputs/parameters . .5 4.1.3 runseq2gene outputs . .8 4.2 gene2pathway . 10 4.2.1 gene2pathway flowchart . 11 4.2.2 gene2pathway test inputs/parameters . 11 4.2.3 gene2pathway test outputs . 12 5 Examples 13 5.1 ChIP-seq data analysis . 13 5.1.1 Map ChIP-seq enriched peaks to genes using runseq2gene .................... 13 5.1.2 Discover enriched GO terms using gene2pathway_test with gene scores . 15 5.1.3 Discover enriched GO terms using Fisher's Exact test without gene scores . 17 5.1.4 Add description for genes . 20 5.2 RNA-seq data analysis . 20 6 R environment session 23 1 Abstract Seq2pathway is a novel computational tool to analyze functional gene-sets (including signaling pathways) using variable next-generation sequencing data[1]. Integral to this tool are the \seq2gene" and \gene2pathway" components in series that infer a quantitative pathway-level profile for each sample. The seq2gene function assigns phenotype-associated significance of genomic regions to gene-level scores, where the significance could be p-values of SNPs or point mutations, protein-binding affinity, or transcriptional expression level. The seq2gene function has the feasibility to assign non-exon regions to a range of neighboring genes besides the nearest one, thus facilitating the study of functional non-coding elements[2]. Then the gene2pathway summarizes gene-level measurements to pathway-level scores, comparing the quantity of significance for gene members within a pathway with those outside a pathway. -

Supporting Online Material
1 2 3 4 5 6 7 Supplementary Information for 8 9 Fractalkine-induced microglial vasoregulation occurs within the retina and is altered early in diabetic 10 retinopathy 11 12 *Samuel A. Mills, *Andrew I. Jobling, *Michael A. Dixon, Bang V. Bui, Kirstan A. Vessey, Joanna A. Phipps, 13 Ursula Greferath, Gene Venables, Vickie H.Y. Wong, Connie H.Y. Wong, Zheng He, Flora Hui, James C. 14 Young, Josh Tonc, Elena Ivanova, Botir T. Sagdullaev, Erica L. Fletcher 15 * Joint first authors 16 17 Corresponding author: 18 Prof. Erica L. Fletcher. Department of Anatomy & Neuroscience. The University of Melbourne, Grattan St, 19 Parkville 3010, Victoria, Australia. 20 Email: [email protected] ; Tel: +61-3-8344-3218; Fax: +61-3-9347-5219 21 22 This PDF file includes: 23 24 Supplementary text 25 Figures S1 to S10 26 Tables S1 to S7 27 Legends for Movies S1 to S2 28 SI References 29 30 Other supplementary materials for this manuscript include the following: 31 32 Movies S1 to S2 33 34 35 36 1 1 Supplementary Information Text 2 Materials and Methods 3 Microglial process movement on retinal vessels 4 Dark agouti rats were anaesthetized, injected intraperitoneally with rhodamine B (Sigma-Aldrich) to label blood 5 vessels and retinal explants established as described in the main text. Retinal microglia were labelled with Iba-1 6 and imaging performed on an inverted confocal microscope (Leica SP5). Baseline images were taken for 10 7 minutes, followed by the addition of PBS (10 minutes) and then either fractalkine or fractalkine + candesartan 8 (10 minutes) using concentrations outlined in the main text. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Patient-Based Cross-Platform Comparison of Oligonucleotide Microarray Expression Profiles
Laboratory Investigation (2005) 85, 1024–1039 & 2005 USCAP, Inc All rights reserved 0023-6837/05 $30.00 www.laboratoryinvestigation.org Patient-based cross-platform comparison of oligonucleotide microarray expression profiles Joerg Schlingemann1,*, Negusse Habtemichael2,*, Carina Ittrich3, Grischa Toedt1, Heidi Kramer1, Markus Hambek4, Rainald Knecht4, Peter Lichter1, Roland Stauber2 and Meinhard Hahn1 1Division of Molecular Genetics, Deutsches Krebsforschungszentrum, Heidelberg, Germany; 2Chemotherapeutisches Forschungsinstitut Georg-Speyer-Haus, Frankfurt am Main, Germany; 3Central Unit Biostatistics, Deutsches Krebsforschungszentrum, Heidelberg, Germany and 4Department of Otorhinolaryngology, Universita¨tsklinik, Johann-Wolfgang-Goethe-Universita¨t Frankfurt, Frankfurt, Germany The comparison of gene expression measurements obtained with different technical approaches is of substantial interest in order to clarify whether interplatform differences may conceal biologically significant information. To address this concern, we analyzed gene expression in a set of head and neck squamous cell carcinoma patients, using both spotted oligonucleotide microarrays made from a large collection of 70-mer probes and commercial arrays produced by in situ synthesis of sets of multiple 25-mer oligonucleotides per gene. Expression measurements were compared for 4425 genes represented on both platforms, which revealed strong correlations between the corresponding data sets. Of note, a global tendency towards smaller absolute ratios was observed when -

Expression Profiling of Ion Channel Genes Predicts Clinical Outcome in Breast Cancer
UCSF UC San Francisco Previously Published Works Title Expression profiling of ion channel genes predicts clinical outcome in breast cancer Permalink https://escholarship.org/uc/item/1zq9j4nw Journal Molecular Cancer, 12(1) ISSN 1476-4598 Authors Ko, Jae-Hong Ko, Eun A Gu, Wanjun et al. Publication Date 2013-09-22 DOI http://dx.doi.org/10.1186/1476-4598-12-106 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Ko et al. Molecular Cancer 2013, 12:106 http://www.molecular-cancer.com/content/12/1/106 RESEARCH Open Access Expression profiling of ion channel genes predicts clinical outcome in breast cancer Jae-Hong Ko1, Eun A Ko2, Wanjun Gu3, Inja Lim1, Hyoweon Bang1* and Tong Zhou4,5* Abstract Background: Ion channels play a critical role in a wide variety of biological processes, including the development of human cancer. However, the overall impact of ion channels on tumorigenicity in breast cancer remains controversial. Methods: We conduct microarray meta-analysis on 280 ion channel genes. We identify candidate ion channels that are implicated in breast cancer based on gene expression profiling. We test the relationship between the expression of ion channel genes and p53 mutation status, ER status, and histological tumor grade in the discovery cohort. A molecular signature consisting of ion channel genes (IC30) is identified by Spearman’s rank correlation test conducted between tumor grade and gene expression. A risk scoring system is developed based on IC30. We test the prognostic power of IC30 in the discovery and seven validation cohorts by both Cox proportional hazard regression and log-rank test. -
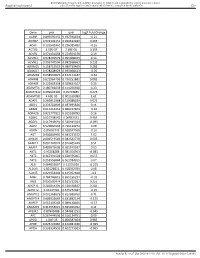
Gene Pval Qval Log2 Fold Change AAMP 0.895690332 0.952598834
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) Gut Gene pval qval Log2 Fold Change AAMP 0.895690332 0.952598834 -0.21 ABI3BP 0.002302151 0.020612283 0.465 ACHE 0.103542461 0.296385483 -0.16 ACTG2 2.99E-07 7.68E-05 3.195 ACVR1 0.071431098 0.224504378 0.19 ACVR1C 0.978209579 0.995008423 0.14 ACVRL1 0.006747504 0.042938663 0.235 ADAM15 0.158715519 0.380719469 0.285 ADAM17 0.978208929 0.995008423 -0.05 ADAM28 0.038932876 0.152174187 -0.62 ADAM8 0.622964796 0.790251882 0.085 ADAM9 0.122003358 0.329623107 0.25 ADAMTS1 0.180766659 0.414256926 0.23 ADAMTS12 0.009902195 0.05703885 0.425 ADAMTS8 4.60E-05 0.001169089 1.61 ADAP1 0.269811968 0.519388039 0.075 ADD1 0.233702809 0.487695826 0.11 ADM2 0.012213453 0.066227879 -0.36 ADRA2B 0.822777921 0.915518785 0.16 AEBP1 0.010738542 0.06035531 0.465 AGGF1 0.117946691 0.320915024 -0.095 AGR2 0.529860903 0.736120272 0.08 AGRN 0.85693743 0.928047568 -0.16 AGT 0.006849995 0.043233572 1.02 AHNAK 0.006519543 0.042542779 0.605 AKAP12 0.001747074 0.016405449 0.51 AKAP2 0.409929603 0.665919397 0.05 AKT1 0.95208288 0.985354963 -0.085 AKT2 0.367391504 0.620376005 0.055 AKT3 0.253556844 0.501934205 0.07 ALB 0.064833867 0.21195036 -0.315 ALDOA 0.83128831 0.918352939 0.08 ALOX5 0.029954404 0.125352668 -0.3 AMH 0.784746815 0.895196237 -0.03 ANG 0.050500474 0.181732067 0.255 ANGPT1 0.281853305 0.538528647 0.285 ANGPT2 0.43147281 0.675272487 -0.15 ANGPTL2 0.001368876 0.013688762 0.71 ANGPTL4 0.686032669 0.831882134 -0.175 ANPEP 0.019103243 0.089148466 -0.57 ANXA2P2 0.412553021 0.665966092 0.11 AP1M2 0.87843088 0.944681253 -0.045 APC 0.267444505 0.516134751 0.09 APOD 1.04E-05 0.000587404 0.985 APOE 0.023722987 0.104981036 -0.395 APOH 0.336334555 0.602273505 -0.065 Sundar R, et al. -

Goblet Cell LRRC26 Regulates BK Channel Activation and Protects Against Colitis in Mice
Goblet cell LRRC26 regulates BK channel activation and protects against colitis in mice Vivian Gonzalez-Pereza,1, Pedro L. Martinez-Espinosaa, Monica Sala-Rabanala, Nikhil Bharadwaja, Xiao-Ming Xiaa, Albert C. Chena,b, David Alvaradoc, Jenny K. Gustafssonc,d,e, Hongzhen Hua, Matthew A. Ciorbab, and Christopher J. Linglea aDepartment of Anesthesiology, Washington University School of Medicine in St. Louis, St. Louis, MO 63110; bMcKelvey School of Engineering, Washington University in St. Louis, St. Louis, MO 63130; cDepartment of Internal Medicine, Division of Gastroenterology, Washington University School of Medicine in St. Louis, St. Louis, MO 63110; dDepartment of Medical Chemistry and Cell Biology, University of Gothenburg, 405 30 Gothenburg, Sweden; and eDepartment of Physiology, University of Gothenburg, 405 30 Gothenburg, Sweden Edited by Richard W. Aldrich, The University of Texas at Austin, Austin, TX, and approved December 21, 2020 (received for review September 16, 2020) Goblet cells (GCs) are specialized cells of the intestinal epithelium Despite this progress, ionic transport in GCs and its implications contributing critically to mucosal homeostasis. One of the func- in GC physiology is a topic that remains poorly understood. tions of GCs is to produce and secrete MUC2, the mucin that forms Here, we address the role of the Ca2+- and voltage-activated K+ the scaffold of the intestinal mucus layer coating the epithelium channel (BK channel) in GCs. and separates the luminal pathogens and commensal microbiota GCs play two primary roles: One related to the maintenance from the host tissues. Although a variety of ion channels and of the mucosal barrier (reviewed in refs.