A Molecular Identification System for Grasses: a Novel Technology
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Triticeae Biodiversity and Conservation, a “Genebanker” View
Czech J. Genet. Plant Breed., 41, 2005 (Special Issue) Triticeae Biodiversity and Conservation, a “Genebanker” View V. H������ Department of Gene Bank, Research Institute of Crop Production, 161 06 Prague-Ruzyně, Czech Republic, e-mail: [email protected] Abstract: Wild Triticeae are important genetic resources for cultivated cereals. While wild and primitive wheats are well preserved, other genera are rather neglected. Most of Triticeae have a large area of distribution, many occupy secondary habitats, or are weedy. However, there are also species with a limited distribution and those need primary attention in conservation. Annuals can be easily stored ex situ as easily as cultivated cereals; peren- nials have their longevity shortened. For successful conservation of genetic diversity one sample of a species is not enough. It is necessary to collect samples from the whole distribution area. Geographically distant popula- tions differ not only morphologically, but can have different spectra of genes. Even within a population there are large differences. For species scattered in distribution or restricted to a certain small area, it is reasonable to consider their in situ conservation. The basic requirement is to protect the locality/ies and to ensure that they are managed for sustainable reproduction of the Triticeae. Basically, this requires maintaining acceptable levels of use by man (grazing and disturbance), acceptable levels of plant competition from other species, and controlling allochtonous invasive species. Localities with in situ conservation require instant monitoring. A detailed docu- mentation (so called passport data) is prerequisite for both ex situ and in situ conservation. The taxonomic system must be conservative, without frequent nomenclatoral changes. -

24. Tribe PANICEAE 黍族 Shu Zu Chen Shouliang (陈守良); Sylvia M
POACEAE 499 hairs, midvein scabrous, apex obtuse, clearly demarcated from mm wide, glabrous, margins spiny-scabrous or loosely ciliate awn; awn 1–1.5 cm; lemma 0.5–1 mm. Anthers ca. 0.3 mm. near base; ligule ca. 0.5 mm. Inflorescence up to 20 cm; spike- Caryopsis terete, narrowly ellipsoid, 1–1.8 mm. lets usually densely arranged, ascending or horizontally spread- ing; rachis scabrous. Spikelets 1.5–2.5 mm (excluding awns); Stream banks, roadsides, other weedy places, on sandy soil. Guangdong, Hainan, Shandong, Taiwan, Yunnan [Bhutan, Cambodia, basal callus 0.1–0.2 mm, obtuse; glumes narrowly lanceolate, India, Indonesia, Laos, Malaysia, Myanmar, Nepal, Philippines, Sri back scaberulous-hirtellous in rather indistinct close rows (most Lanka, Thailand, Vietnam; Africa (probably introduced), Australia obvious toward lemma base), midvein pectinate-ciliolate, apex (Queensland)]. abruptly acute, clearly demarcated from awn; awn 0.5–1.5 cm. Anthers ca. 0.3 mm. Caryopsis terete, narrowly ellipsoid, ca. 3. Perotis hordeiformis Nees in Hooker & Arnott, Bot. Beech- 1.5 mm. Fl. and fr. summer and autumn. 2n = 40. ey Voy. 248. 1838. Sandy places, along seashores. Guangdong, Hebei, Jiangsu, 麦穗茅根 mai sui mao gen Yunnan [India, Indonesia, Malaysia, Nepal, Myanmar, Pakistan, Sri Lanka, Thailand]. Perotis chinensis Gandoger. This species is very close to Perotis indica and is sometimes in- Annual or short-lived perennial. Culms loosely tufted, cluded within it. No single character by itself is reliable for separating erect or decumbent at base, 25–40 cm tall. Leaf sheaths gla- the two, but the combination of characters given in the key will usually brous; leaf blades lanceolate to narrowly ovate, 2–4 cm, 4–7 suffice. -

Breeding System Diversification and Evolution in American Poa Supersect. Homalopoa (Poaceae: Poeae: Poinae)
Annals of Botany Page 1 of 23 doi:10.1093/aob/mcw108, available online at www.aob.oxfordjournals.org Breeding system diversification and evolution in American Poa supersect. Homalopoa (Poaceae: Poeae: Poinae) Liliana M. Giussani1,*, Lynn J. Gillespie2, M. Amalia Scataglini1,Marıa A. Negritto3, Ana M. Anton4 and Robert J. Soreng5 1Instituto de Botanica Darwinion, San Isidro, Buenos Aires, Argentina, 2Research and Collections Division, Canadian Museum of Nature, Ottawa, Ontario, Canada, 3Universidad de Magdalena, Santa Marta, Colombia, 4Instituto Multidisciplinario de Biologıa Vegetal (IMBIV), CONICET-UNC, Cordoba, Argentina and 5Department of Botany, Smithsonian Institution, Washington, DC, USA *For correspondence. E-mail [email protected] Received: 11 December 2015 Returned for revision: 18 February 2016 Accepted: 18 March 2016 Downloaded from Background and Aims Poa subgenus Poa supersect. Homalopoa has diversified extensively in the Americas. Over half of the species in the supersection are diclinous; most of these are from the New World, while a few are from South-East Asia. Diclinism in Homalopoa can be divided into three main types: gynomonoecism, gynodioe- cism and dioecism. Here the sampling of species of New World Homalopoa is expanded to date its origin and diver- sification in North and South America and examine the evolution and origin of the breeding system diversity. Methods A total of 124 specimens were included in the matrix, of which 89 are species of Poa supersect. http://aob.oxfordjournals.org/ Homalopoa sections Acutifoliae, Anthochloa, Brizoides, Dasypoa, Dioicopoa, Dissanthelium, Homalopoa sensu lato (s.l.), Madropoa and Tovarochloa, and the informal Punapoa group. Bayesian and parsimony analyses were conducted on the data sets based on four markers: the nuclear ribosomal internal tanscribed spacer (ITS) and exter- nal transcribed spacer (ETS), and plastid trnT-L and trnL-F. -

Registration of the Triticeae-CAP Spring Wheat Nested Association Mapping Population
Published February 28, 2019 JOURNAL OF PLANT REGISTRATIONS MAPPING POPULATION Registration of the Triticeae-CAP Spring Wheat Nested Association Mapping Population N. K. Blake, M. Pumphrey, K. Glover, S. Chao, K. Jordan, J.-L. Jannick, E. A. Akhunov, J. Dubcovsky, H. Bockelman, and L. E. Talbert* Abstract andrace accessions of wheat (Triticum aestivum The Triticeae-CAP spring wheat nested association mapping L.) are a potentially important resource for superior population (Reg. No. MP-10, NSL 527060 MAP) consisting of genes to improve modern wheat cultivars. The landrace recombinant inbred line (RIL) populations derived from 32 Laccessions themselves are typically inferior to modern cultivars spring wheat (Triticum aestivum L.) accessions each crossed to for both agronomic and quality characteristics. This is particu- a common spring wheat parent, ‘Berkut’, has been released. larly true when landraces from one region are grown in a differ- The spring wheat accessions consisted of 29 landraces and ent locale in that important alleles for adaptation to biotic and three cultivars. Each population consists of approximately 75 abiotic factors are lacking. A challenge for breeders is that supe- lines for a total of 2325 RILs (Reg. Nos. GSTR No. 14701–GSTR 17133). The RILs have all been genotyped with the Illumina rior alleles are difficult to detect in a background of undesir- wheat iSelect 90K single nucleotide polymorphism array able alleles that typify the landraces. The increased efficiency of using the Infinium assay method and through genotype-by- modern genotyping capabilities has provided an opportunity to sequencing. This nested association mapping population identify superior alleles from nonadapted germplasm for incor- provides a genotyped germplasm resource for the wheat poration into elite breeding material. -
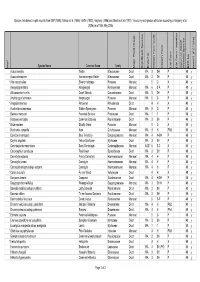
BFS048 Site Species List
Species lists based on plot records from DEP (1996), Gibson et al. (1994), Griffin (1993), Keighery (1996) and Weston et al. (1992). Taxonomy and species attributes according to Keighery et al. (2006) as of 16th May 2005. Species Name Common Name Family Major Plant Group Significant Species Endemic Growth Form Code Growth Form Life Form Life Form - aquatics Common SSCP Wetland Species BFS No kens01 (FCT23a) Wd? Acacia sessilis Wattle Mimosaceae Dicot WA 3 SH P 48 y Acacia stenoptera Narrow-winged Wattle Mimosaceae Dicot WA 3 SH P 48 y * Aira caryophyllea Silvery Hairgrass Poaceae Monocot 5 G A 48 y Alexgeorgea nitens Alexgeorgea Restionaceae Monocot WA 6 S-R P 48 y Allocasuarina humilis Dwarf Sheoak Casuarinaceae Dicot WA 3 SH P 48 y Amphipogon turbinatus Amphipogon Poaceae Monocot WA 5 G P 48 y * Anagallis arvensis Pimpernel Primulaceae Dicot 4 H A 48 y Austrostipa compressa Golden Speargrass Poaceae Monocot WA 5 G P 48 y Banksia menziesii Firewood Banksia Proteaceae Dicot WA 1 T P 48 y Bossiaea eriocarpa Common Bossiaea Papilionaceae Dicot WA 3 SH P 48 y * Briza maxima Blowfly Grass Poaceae Monocot 5 G A 48 y Burchardia congesta Kara Colchicaceae Monocot WA 4 H PAB 48 y Calectasia narragara Blue Tinsel Lily Dasypogonaceae Monocot WA 4 H-SH P 48 y Calytrix angulata Yellow Starflower Myrtaceae Dicot WA 3 SH P 48 y Centrolepis drummondiana Sand Centrolepis Centrolepidaceae Monocot AUST 6 S-C A 48 y Conostephium pendulum Pearlflower Epacridaceae Dicot WA 3 SH P 48 y Conostylis aculeata Prickly Conostylis Haemodoraceae Monocot WA 4 H P 48 y Conostylis juncea Conostylis Haemodoraceae Monocot WA 4 H P 48 y Conostylis setigera subsp. -

Phylogeny and Subfamilial Classification of the Grasses (Poaceae) Author(S): Grass Phylogeny Working Group, Nigel P
Phylogeny and Subfamilial Classification of the Grasses (Poaceae) Author(s): Grass Phylogeny Working Group, Nigel P. Barker, Lynn G. Clark, Jerrold I. Davis, Melvin R. Duvall, Gerald F. Guala, Catherine Hsiao, Elizabeth A. Kellogg, H. Peter Linder Source: Annals of the Missouri Botanical Garden, Vol. 88, No. 3 (Summer, 2001), pp. 373-457 Published by: Missouri Botanical Garden Press Stable URL: http://www.jstor.org/stable/3298585 Accessed: 06/10/2008 11:05 Your use of the JSTOR archive indicates your acceptance of JSTOR's Terms and Conditions of Use, available at http://www.jstor.org/page/info/about/policies/terms.jsp. JSTOR's Terms and Conditions of Use provides, in part, that unless you have obtained prior permission, you may not download an entire issue of a journal or multiple copies of articles, and you may use content in the JSTOR archive only for your personal, non-commercial use. Please contact the publisher regarding any further use of this work. Publisher contact information may be obtained at http://www.jstor.org/action/showPublisher?publisherCode=mobot. Each copy of any part of a JSTOR transmission must contain the same copyright notice that appears on the screen or printed page of such transmission. JSTOR is a not-for-profit organization founded in 1995 to build trusted digital archives for scholarship. We work with the scholarly community to preserve their work and the materials they rely upon, and to build a common research platform that promotes the discovery and use of these resources. For more information about JSTOR, please contact [email protected]. -

The Genera of Bambusoideae (Gramineae) in the Southeastern United States Gordon C
Eastern Illinois University The Keep Faculty Research & Creative Activity Biological Sciences January 1988 The genera of Bambusoideae (Gramineae) in the southeastern United States Gordon C. Tucker Eastern Illinois University, [email protected] Follow this and additional works at: http://thekeep.eiu.edu/bio_fac Part of the Biology Commons Recommended Citation Tucker, Gordon C., "The eg nera of Bambusoideae (Gramineae) in the southeastern United States" (1988). Faculty Research & Creative Activity. 181. http://thekeep.eiu.edu/bio_fac/181 This Article is brought to you for free and open access by the Biological Sciences at The Keep. It has been accepted for inclusion in Faculty Research & Creative Activity by an authorized administrator of The Keep. For more information, please contact [email protected]. TUCKER, BAMBUSOIDEAE 239 THE GENERA OF BAMBUSOIDEAE (GRAMINEAE) IN THE SOUTHEASTERN UNITED STATESu GoRDON C. T ucKER3 Subfamily BAMBUSOIDEAE Ascherson & Graebner, Synop. Mitteleurop. Fl. 2: 769. 1902. Perennial or annual herbs or woody plants of tropical or temperate forests and wetlands. Rhizomes present or lacking. Stems erect or decumbent (some times rooting at the lower nodes); nodes glabrous, pubescent, or puberulent. Leaves several to many, glabrous to sparsely pubescent (microhairs bicellular); leaf sheaths about as long as the blades, open for over tf2 their length, glabrous; ligules wider than long, entire or fimbriate; blades petiolate or sessile, elliptic to linear, acute to acuminate, the primary veins parallel to-or forming an angle of 5-10• wi th-the midvein, transverse veinlets numerous, usually con spicuous, giving leaf surface a tessellate appearance; chlorenchyma not radiate (i.e., non-kranz; photosynthetic pathway C.,). -

By H.D.V. PRENDERGAST a Thesis Submitted for the Degree of Doctor of Philosophy of the Australian National University. January 1
STRUCTURAL, BIOCHEMICAL AND GEOGRAPHICAL RELATIONSHIPS IN AUSTRALIAN c4 GRASSES (POACEAE) • by H.D.V. PRENDERGAST A thesis submitted for the degree of Doctor of Philosophy of the Australian National University. January 1987. Canberra, Australia. i STATEMENT This thesis describes my own work which included collaboration with Dr N .. E. Stone (Taxonomy Unit, R .. S .. B.S .. ), whose expertise in enzyme assays enabled me to obtain comparative information on enzyme activities reported in Chapters 3, 5 and 7; and with Mr M.. Lazarides (Australian National Herbarium, c .. s .. r .. R .. O .. ), whose as yet unpublished taxonomic views on Eragrostis form the basis of some of the discussion in Chapter 3. ii This thesis describes the results of research work carried out in the Taxonomy Unit, Research School of Biological Sciences, The Australian National University during the tenure of an A.N.U. Postgraduate Scholarship. iii ACKNOWLEDGEMENTS My time in the Taxonomy Unit has been a happy one: I could not have asked for better supervision for my project or for a more congenial atmosphere in which to work. To Dr. Paul Hattersley, for his help, advice, encouragement and friendship, I owe a lot more than can be said in just a few words: but, Paul, thanks very much! To Mr. Les Watson I owe as much for his own support and guidance, and for many discussions on things often psittacaceous as well as graminaceous! Dr. Nancy Stone was a kind teacher in many days of enzyme assays and Chris Frylink a great help and friend both in and out of the lab •• Further thanks go to Mike Lazarides (Australian National Herbarium, c.s.I.R.O.) for identifying many grass specimens and for unpublished data on infrageneric groups in Eragrostis; Dr. -

Urochloa Subquadripara (Poaceae: Paniceae) New to Texas and a Key to Urochloa of Texas
Hatch, S.L. 2010. Urochloa subquadripara (Poaceae: Paniceae) new to Texas and a key to Urochloa of Texas. Phytoneuron 2010-8: 1-4. (8 April) UROCHLOA SUBQUADRIPARA (POACEAE: PANICEAE) NEW TO TEXAS AND A KEY TO UROCHLOA OF TEXAS Stephan L. Hatch S.M. Tracy Herbarium (TAES) Department of Ecosystem Science and Management Texas A&M University College Station, TX 77843-2138, U.S.A. [email protected] ABSTRACT Urochloa subquadripara is reported as introduced into Texas. A key to separate the 13 species of Urochloa in Texas is presented along with an image of the newly reported species. KEY WORDS : Poaceae, Urochloa , Texas, introduced, invasive plant Urochloa P. Beauv. is primarily a grass genus of Old World origin. Thirteen of the estimated 100 species (Wipff & Thompson 2003) worldwide occur in Texas. This genus was separated from closely related or similar Paniceae by Wipff et al. in 1993. Urochloa (Wipff & Thompson 2003) is described as having terminal and axilliary panicle inflorescences with 2 to several spicate primary unilateral branches. Spikelets are solitary, paired, or in triplets and occur in 1–2 (4) rows per primary branch. With 2 florets per spikelet, the upper floret is fertile, indurate and rugose to verrucose, the lower floret sterile or staminate. A key to three Urochloa species was published by Wipff et al. (1993). Eight of the Texas taxa are introduced (five invasive) and five are native to North America. The introduced taxa are native to tropical or subtropical regions of the world and their points of introduction appear to be from the coast or south Texas and following a period of adaptation move inland and/or to the north. -

Evolution in Sedges (Carex, Cyperaceae)
Evolution in sedges (Carex, Cyperaceae) A. A. REZNICEK University of Michigan Herbarium, North University Building, Ann Arbor, MI 48/09, U.S.A. Received January 2, 1990 REZNICEK,A. A. 1990. Evolution in sedges (Carex, Cyperaceae). Can. J. Bot. 68: 1409-1432. Carex is the largest and most widespread genus of Cyperaceae, but evolutionary relationships within it are poorly under- stood. Subgenus Primocarex was generally thought to be artificial and derived from diverse multispicate species. Relation- ships of rachilla-bearing species of subgenus Primocarex, however, were disputed, with some authors suggesting derivation from other genera, and others believing them to be primitive. Subgenus Indocarex, with compounded inflorescence units, was thought to be primitive, with subgenera Carex and Vignea reduced and derived. However, occurrence of rachillas is not confined to a few unispicate species, as previously thought, but is widespread. The often suggested connection between Uncinia and unispicate Carex is shown, based on rachilla morphology, to be founded on incorrect interpretation OF homology. Uncinia kingii, the alleged connecting link, is, in fact, a Carex. Unispicate Carex without close multispicate relatives probably originated from independent, ancient reductions of primitive, rachilla-bearing, multispicate Carex. The highly compounded inflorescences occumng in subgenus Vignea are hypothesized to represent a primitive state in Carex, and the more specialized inflorescences in subgenus Carex derived from inflorescences of this type. The relationships of subgenus Indocurex, with its unique perigynium-like inflorescence prophylls, remain unclear. REZNICEK,A. A. 1990. Evolution in sedges (Carex, Cyperaceae). Can. J. Bot. 68 : 1409-1432. Le Carex est le genre le plus irilportant et le plus rCpandu des Cyperaceae, mais les affinites Cvolutives a I'intCrieur de ce genre sont ma1 connues. -

Poaceae: Pooideae) Based on Plastid and Nuclear DNA Sequences
d i v e r s i t y , p h y l o g e n y , a n d e v o l u t i o n i n t h e monocotyledons e d i t e d b y s e b e r g , p e t e r s e n , b a r f o d & d a v i s a a r h u s u n i v e r s i t y p r e s s , d e n m a r k , 2 0 1 0 Phylogenetics of Stipeae (Poaceae: Pooideae) Based on Plastid and Nuclear DNA Sequences Konstantin Romaschenko,1 Paul M. Peterson,2 Robert J. Soreng,2 Núria Garcia-Jacas,3 and Alfonso Susanna3 1M. G. Kholodny Institute of Botany, Tereshchenkovska 2, 01601 Kiev, Ukraine 2Smithsonian Institution, Department of Botany MRC-166, National Museum of Natural History, P.O. Box 37012, Washington, District of Columbia 20013-7012 USA. 3Laboratory of Molecular Systematics, Botanic Institute of Barcelona (CSIC-ICUB), Pg. del Migdia, s.n., E08038 Barcelona, Spain Author for correspondence ([email protected]) Abstract—The Stipeae tribe is a group of 400−600 grass species of worldwide distribution that are currently placed in 21 genera. The ‘needlegrasses’ are char- acterized by having single-flowered spikelets and stout, terminally-awned lem- mas. We conducted a molecular phylogenetic study of the Stipeae (including all genera except Anemanthele) using a total of 94 species (nine species were used as outgroups) based on five plastid DNA regions (trnK-5’matK, matK, trnHGUG-psbA, trnL5’-trnF, and ndhF) and a single nuclear DNA region (ITS). -

Grasses of the Texas Hill Country: Vegetative Key and Descriptions
Hagenbuch, K.W. and D.E. Lemke. 2015. Grasses of the Texas Hill Country: Vegetative key and descriptions. Phytoneuron 2015-4: 1–93. Published 7 January 2015. ISSN 2153 733X GRASSES OF THE TEXAS HILL COUNTRY: VEGETATIVE KEY AND DESCRIPTIONS KARL W. HAGENBUCH Department of Biological Sciences San Antonio College 1300 San Pedro Avenue San Antonio, Texas 78212-4299 [email protected] DAVID E. LEMKE Department of Biology Texas State University 601 University Drive San Marcos, Texas 78666-4684 [email protected] ABSTRACT A key and a set of descriptions, based solely on vegetative characteristics, is provided for the identification of 66 genera and 160 grass species, both native and naturalized, of the Texas Hill Country. The principal characters used (features of longevity, growth form, roots, rhizomes and stolons, culms, leaf sheaths, collars, auricles, ligules, leaf blades, vernation, vestiture, and habitat) are discussed and illustrated. This treatment should prove useful at times when reproductive material is not available. Because of its size and variation in environmental conditions, Texas provides habitat for well over 700 species of grasses (Shaw 2012). For identification purposes, the works of Correll and Johnston (1970); Gould (1975) and, more recently, Shaw (2012) treat Texas grasses in their entirety. In addition to these comprehensive works, regional taxonomic treatments have been done for the grasses of the Cross Timbers and Prairies (Hignight et al. 1988), the South Texas Brush Country (Lonard 1993; Everitt et al. 2011), the Gulf Prairies and Marshes (Hatch et al. 1999), and the Trans-Pecos (Powell 1994) natural regions. In these, as well as in numerous other manuals and keys, accurate identification of grass species depends on the availability of reproductive material.