Pontibacillus Yanchengensis Sp. Nov., a Moderately Halophilic Bacterium Isolated from Salt Field Soil
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Bacillus Crassostreae Sp. Nov., Isolated from an Oyster (Crassostrea Hongkongensis)
International Journal of Systematic and Evolutionary Microbiology (2015), 65, 1561–1566 DOI 10.1099/ijs.0.000139 Bacillus crassostreae sp. nov., isolated from an oyster (Crassostrea hongkongensis) Jin-Hua Chen,1,2 Xiang-Rong Tian,2 Ying Ruan,1 Ling-Ling Yang,3 Ze-Qiang He,2 Shu-Kun Tang,3 Wen-Jun Li,3 Huazhong Shi4 and Yi-Guang Chen2 Correspondence 1Pre-National Laboratory for Crop Germplasm Innovation and Resource Utilization, Yi-Guang Chen Hunan Agricultural University, 410128 Changsha, PR China [email protected] 2College of Biology and Environmental Sciences, Jishou University, 416000 Jishou, PR China 3The Key Laboratory for Microbial Resources of the Ministry of Education, Yunnan Institute of Microbiology, Yunnan University, 650091 Kunming, PR China 4Department of Chemistry and Biochemistry, Texas Tech University, Lubbock, TX 79409, USA A novel Gram-stain-positive, motile, catalase- and oxidase-positive, endospore-forming, facultatively anaerobic rod, designated strain JSM 100118T, was isolated from an oyster (Crassostrea hongkongensis) collected from the tidal flat of Naozhou Island in the South China Sea. Strain JSM 100118T was able to grow with 0–13 % (w/v) NaCl (optimum 2–5 %), at pH 5.5–10.0 (optimum pH 7.5) and at 5–50 6C (optimum 30–35 6C). The cell-wall peptidoglycan contained meso-diaminopimelic acid as the diagnostic diamino acid. The predominant respiratory quinone was menaquinone-7 and the major cellular fatty acids were anteiso-C15 : 0, iso-C15 : 0,C16 : 0 and C16 : 1v11c. The polar lipids consisted of diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylglycerol, an unknown glycolipid and an unknown phospholipid. The genomic DNA G+C content was 35.9 mol%. -

Desulfuribacillus Alkaliarsenatis Gen. Nov. Sp. Nov., a Deep-Lineage
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by PubMed Central Extremophiles (2012) 16:597–605 DOI 10.1007/s00792-012-0459-7 ORIGINAL PAPER Desulfuribacillus alkaliarsenatis gen. nov. sp. nov., a deep-lineage, obligately anaerobic, dissimilatory sulfur and arsenate-reducing, haloalkaliphilic representative of the order Bacillales from soda lakes D. Y. Sorokin • T. P. Tourova • M. V. Sukhacheva • G. Muyzer Received: 10 February 2012 / Accepted: 3 May 2012 / Published online: 24 May 2012 Ó The Author(s) 2012. This article is published with open access at Springerlink.com Abstract An anaerobic enrichment culture inoculated possible within a pH range from 9 to 10.5 (optimum at pH with a sample of sediments from soda lakes of the Kulunda 10) and a salt concentration at pH 10 from 0.2 to 2 M total Steppe with elemental sulfur as electron acceptor and for- Na? (optimum at 0.6 M). According to the phylogenetic mate as electron donor at pH 10 and moderate salinity analysis, strain AHT28 represents a deep independent inoculated with sediments from soda lakes in Kulunda lineage within the order Bacillales with a maximum of Steppe (Altai, Russia) resulted in the domination of a 90 % 16S rRNA gene similarity to its closest cultured Gram-positive, spore-forming bacterium strain AHT28. representatives. On the basis of its distinct phenotype and The isolate is an obligate anaerobe capable of respiratory phylogeny, the novel haloalkaliphilic anaerobe is suggested growth using elemental sulfur, thiosulfate (incomplete as a new genus and species, Desulfuribacillus alkaliar- T T reduction) and arsenate as electron acceptor with H2, for- senatis (type strain AHT28 = DSM24608 = UNIQEM mate, pyruvate and lactate as electron donor. -

Previously Uncultured Marine Bacteria Linked to Novel Alkaloid Production
UC San Diego UC San Diego Previously Published Works Title Previously Uncultured Marine Bacteria Linked to Novel Alkaloid Production. Permalink https://escholarship.org/uc/item/6263h6gw Journal Chemistry & biology, 22(9) ISSN 1074-5521 Authors Choi, Eun Ju Nam, Sang-Jip Paul, Lauren et al. Publication Date 2015-09-01 DOI 10.1016/j.chembiol.2015.07.014 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Resource Previously Uncultured Marine Bacteria Linked to Novel Alkaloid Production Graphical Abstract Authors Eun Ju Choi, Sang-Jip Nam, Lauren Paul, ..., Christopher A. Kauffman, Paul R. Jensen, William Fenical Correspondence [email protected] In Brief Choi et al. illustrate that low-nutrient media and long incubation times lead to the isolation of rare, previously uncultured marine bacteria that produce new antibacterial metabolites. Their work demonstrates that unique marine bacteria are easier to cultivate than previously suggested. Highlights Accession Numbers d Simple methods allow the isolation of rare, previously JN703500 KJ572269 uncultured marine bacteria JN703501 KJ572270 JN703502 KJ572271 d Previously uncultured marine bacteria produce new JN703503 KJ572272 antibacterial metabolites JN368460 KJ572273 JN368461 KJ572274 KJ572262 KJ572275 KJ572263 KJ572264 KJ572265 KJ572266 KJ572267 KJ572268 Choi et al., 2015, Chemistry & Biology 22, 1–10 September 17, 2015 ª2015 Elsevier Ltd All rights reserved http://dx.doi.org/10.1016/j.chembiol.2015.07.014 Please cite this article in press as: Choi et al., Previously Uncultured Marine Bacteria Linked to Novel Alkaloid Production, Chemistry & Biology (2015), http://dx.doi.org/10.1016/j.chembiol.2015.07.014 Chemistry & Biology Resource Previously Uncultured Marine Bacteria Linked to Novel Alkaloid Production Eun Ju Choi,1,2,3 Sang-Jip Nam,1,2,4 Lauren Paul,1 Deanna Beatty,1 Christopher A. -

A Novel Α-Amylase from Marine Bacterium Pontibacillus Amyz1
Florida International University FIU Digital Commons Department of Chemistry and Biochemistry College of Arts, Sciences & Education 4-23-2019 AmyZ1: a novel α-amylase from marine bacterium Pontibacillus sp. ZY with high activity toward raw starches Wei Fang Saisai Xue Pengjun Deng Xuecheng Zhang Xiaotang Wang See next page for additional authors Follow this and additional works at: https://digitalcommons.fiu.edu/chemistry_fac Part of the Chemistry Commons This work is brought to you for free and open access by the College of Arts, Sciences & Education at FIU Digital Commons. It has been accepted for inclusion in Department of Chemistry and Biochemistry by an authorized administrator of FIU Digital Commons. For more information, please contact [email protected]. Authors Wei Fang, Saisai Xue, Pengjun Deng, Xuecheng Zhang, Xiaotang Wang, Yazhong Xiao, and Zemin Fang Fang et al. Biotechnol Biofuels (2019) 12:95 https://doi.org/10.1186/s13068-019-1432-9 Biotechnology for Biofuels RESEARCH Open Access AmyZ1: a novel α-amylase from marine bacterium Pontibacillus sp. ZY with high activity toward raw starches Wei Fang1,2,3† , Saisai Xue1,2,3†, Pengjun Deng1,2,3, Xuecheng Zhang1,2,3, Xiaotang Wang4, Yazhong Xiao1,2,3* and Zemin Fang1,2,3* Abstract Background: Starch is an inexpensive and renewable raw material for numerous industrial applications. However, most starch-based products are not cost-efcient due to high-energy input needed in traditional enzymatic starch conversion processes. Therefore, α-amylase with high efciency to directly hydrolyze high concentration raw starches at a relatively lower temperature will have a profound impact on the efcient application of starch. -

Increases in Soil Respiration Following Labile Carbon Additions Linked to Rapid Shifts in Soil Microbial Community Composition
Biogeochemistry D O I 10.1007/S10533-006-9065-Z ORIGINAL PAPER Increases in soil respiration following labile carbon additions linked to rapid shifts in soil microbial community composition Cory C. Cleveland • Diana R. Nemergnt Steven K. Schmidt • Alan R. Townsend Received: 16 June 2006 / Accepted: 10 November 2006 © Springer Science+Business Media B.V. 2006 Abstract Organic matter decomposition and soil determined by constructing clone libraries of CO 2 efflux are both mediated by soil microorgan small-subunit ribosomal RNA genes (SSU rRNA) isms, but the potential effects of temporal varia extracted from the soil at the end of the incubation tions in microbial community composition are not experiment. In contrast to the subtle effects of considered in most analytical models of these adding water alone, additions of DOM caused a two important processes. However, inconsistent rapid and large increase in soil CO2 flux. DOM- relationships between rates of heterotrophic soil stimulated CO2 fluxes also coincided with pro respiration and abiotic factors, including temper found shifts in the abundance of certain members ature and moisture, suggest that microbial com of the soil microbial community. Our results munity composition may be an important regulator suggest that natural DOM inputs may drive high of soil organic matter (SOM) decomposition and rates of soil respiration by stimulating an opportu CO 2 efflux. We performed a short-term (12-h) nistic subset of the soil bacterial community, laboratory incubation experiment using tropical particularly members of the Gammaproteobacte- rain forest soil amended with either water (as a ria and Firmicutes groups. Our experiment indi control) or dissolved organic matter (DOM) cates that variations in microbial community leached from native plant litter, and analyzed the composition may influence SOM decomposition effects of the treatments on soil respiration and and soil respiration rates, and emphasizes the need microbial community composition. -

Thermolongibacillus Cihan Et Al
Genus Firmicutes/Bacilli/Bacillales/Bacillaceae/ Thermolongibacillus Cihan et al. (2014)VP .......................................................................................................................................................................................... Arzu Coleri Cihan, Department of Biology, Faculty of Science, Ankara University, Ankara, Turkey Kivanc Bilecen and Cumhur Cokmus, Department of Molecular Biology & Genetics, Faculty of Agriculture & Natural Sciences, Konya Food & Agriculture University, Konya, Turkey Ther.mo.lon.gi.ba.cil’lus. Gr. adj. thermos hot; L. adj. Type species: Thermolongibacillus altinsuensis E265T, longus long; L. dim. n. bacillus small rod; N.L. masc. n. DSM 24979T, NCIMB 14850T Cihan et al. (2014)VP. .................................................................................. Thermolongibacillus long thermophilic rod. Thermolongibacillus is a genus in the phylum Fir- Gram-positive, motile rods, occurring singly, in pairs, or micutes,classBacilli, order Bacillales, and the family in long straight or slightly curved chains. Moderate alka- Bacillaceae. There are two species in the genus Thermo- lophile, growing in a pH range of 5.0–11.0; thermophile, longibacillus, T. altinsuensis and T. kozakliensis, isolated growing in a temperature range of 40–70∘C; halophile, from sediment and soil samples in different ther- tolerating up to 5.0% (w/v) NaCl. Catalase-weakly positive, mal hot springs, respectively. Members of this genus chemoorganotroph, grow aerobically, but not under anaer- are thermophilic (40–70∘C), halophilic (0–5.0% obic conditions. Young cells are 0.6–1.1 μm in width and NaCl), alkalophilic (pH 5.0–11.0), endospore form- 3.0–8.0 μm in length; cells in stationary and death phases ing, Gram-positive, aerobic, motile, straight rods. are 0.6–1.2 μm in width and 9.0–35.0 μm in length. -

Isolation and Diversity of Sediment Bacteria in The
bioRxiv preprint doi: https://doi.org/10.1101/638304; this version posted May 14, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 Isolation and Diversity of Sediment Bacteria in the 2 Hypersaline Aiding Lake, China 3 4 Tong-Wei Guan, Yi-Jin Lin, Meng-Ying Ou, Ke-Bao Chen 5 6 7 Institute of Microbiology, Xihua University, Chengdu 610039, P. R. China. 8 9 Author for correspondence: 10 Tong-Wei Guan 11 Tel/Fax: +86 028 87720552 12 E-mail: [email protected] 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 bioRxiv preprint doi: https://doi.org/10.1101/638304; this version posted May 14, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 29 Abstract A total of 343 bacteria from sediment samples of Aiding Lake, China, were isolated using 30 nine different media with 5% or 15% (w/v) NaCl. The number of species and genera of bacteria recovered 31 from the different media significantly varied, indicating the need to optimize the isolation conditions. 32 The results showed an unexpected level of bacterial diversity, with four phyla (Firmicutes, 33 Actinobacteria, Proteobacteria, and Rhodothermaeota), fourteen orders (Actinopolysporales, 34 Alteromonadales, Bacillales, Balneolales, Chromatiales, Glycomycetales, Jiangellales, Micrococcales, 35 Micromonosporales, Oceanospirillales, Pseudonocardiales, Rhizobiales, Streptomycetales, and 36 Streptosporangiales), including 17 families, 41 genera, and 71 species. -

Isolation and Diversity of Sediment Bacteria in The
bioRxiv preprint doi: https://doi.org/10.1101/638304; this version posted May 14, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 Isolation and Diversity of Sediment Bacteria in the 2 Hypersaline Aiding Lake, China 3 4 Tong-Wei Guan, Yi-Jin Lin, Meng-Ying Ou, Ke-Bao Chen 5 6 7 Institute of Microbiology, Xihua University, Chengdu 610039, P. R. China. 8 9 Author for correspondence: 10 Tong-Wei Guan 11 Tel/Fax: +86 028 87720552 12 E-mail: [email protected] 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 bioRxiv preprint doi: https://doi.org/10.1101/638304; this version posted May 14, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 29 Abstract A total of 343 bacteria from sediment samples of Aiding Lake, China, were isolated using 30 nine different media with 5% or 15% (w/v) NaCl. The number of species and genera of bacteria recovered 31 from the different media significantly varied, indicating the need to optimize the isolation conditions. 32 The results showed an unexpected level of bacterial diversity, with four phyla (Firmicutes, 33 Actinobacteria, Proteobacteria, and Rhodothermaeota), fourteen orders (Actinopolysporales, 34 Alteromonadales, Bacillales, Balneolales, Chromatiales, Glycomycetales, Jiangellales, Micrococcales, 35 Micromonosporales, Oceanospirillales, Pseudonocardiales, Rhizobiales, Streptomycetales, and 36 Streptosporangiales), including 17 families, 41 genera, and 71 species. -
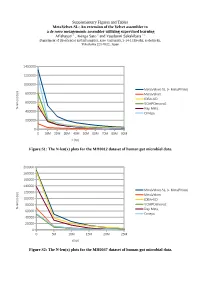
Supplementary Figures and Tables Metavelvet-SL
Supplementary Figures and Tables MetaVelvet-SL: An extension of the Velvet assembler to a de novo metagenomic assembler utilizing supervised learning Afiahayati 1 , Kengo Sato 1 and Yasubumi Sakakibara 1∗ Department of Biosciences and Informatics, Keio University, 3-14-1 Hiyoshi, Kohoku-ku, Yokohama 223-8522, Japan 1400000 1200000 1000000 MetaVelvet-SL (+ MetaPhlAn) ) p 800000 b MetaVelvet ( ) x ( IDBA-UD n 600000 e SOAPDenovo2 l - N Ray Meta 400000 Omega 200000 0 0 10M 20M 30M 40M 50M 60M 70M 80M 90M x (bp) Figure S1: The N-len(x) plots for the MH0012 dataset of human gut microbial data. 200000 180000 160000 140000 MetaVelvet-SL (+ MetaPhlAn) ) 120000 p b MetaVelvet ( ) 100000 x IDBA-UD ( n e l 80000 SOAPDenovo2 - N Ray Meta 60000 Omega 40000 20000 0 0 5M 10M 15M 20M 25M x(bp) Figure S2: The N-len(x) plots for the MH0047 dataset of human gut microbial data. 600000 500000 400000 MetaVelvet-SL (+ MetaPhlAn) ) p b MetaVelvet ( ) 300000 x IDBA-UD ( n e l SOAPDenovo2 - N 200000 Ray Meta Omega 100000 0 0 10M 20M 30M 40M 50M 60M 70M 80M 90M x (bp) Figure S3: The N-len(x) plots for the SRS017227 dataset of human gut microbial data. 800000 700000 600000 500000 MetaVelvet-SL (+ MetaPhlAn) ) p b MetaVelvet ( ) 400000 x IDBA-UD ( n e l SOAPDenovo2 - 300000 N Ray Meta 200000 Omega 100000 0 0 5M 10M 15M 20M 25M 30M 35M 40M 45M x (bp) Figure S4: The N-len(x) plots for the SRS018661 dataset of human gut microbial data. Formula to identify unique nodes in Velvet (Zerbino and Birney, 2008) ̄x2 ρ2− log2 2 F ( ̄x ,n,ρ )= +n 2 2 where ̄x = the coverage of node ρ = expected coverage of subgraph n = the length of node A node is “unique”, if its F > 5 . -

Reorganising the Order Bacillales Through Phylogenomics
Systematic and Applied Microbiology 42 (2019) 178–189 Contents lists available at ScienceDirect Systematic and Applied Microbiology jou rnal homepage: http://www.elsevier.com/locate/syapm Reorganising the order Bacillales through phylogenomics a,∗ b c Pieter De Maayer , Habibu Aliyu , Don A. Cowan a School of Molecular & Cell Biology, Faculty of Science, University of the Witwatersrand, South Africa b Technical Biology, Institute of Process Engineering in Life Sciences, Karlsruhe Institute of Technology, Germany c Centre for Microbial Ecology and Genomics, University of Pretoria, South Africa a r t i c l e i n f o a b s t r a c t Article history: Bacterial classification at higher taxonomic ranks such as the order and family levels is currently reliant Received 7 August 2018 on phylogenetic analysis of 16S rRNA and the presence of shared phenotypic characteristics. However, Received in revised form these may not be reflective of the true genotypic and phenotypic relationships of taxa. This is evident in 21 September 2018 the order Bacillales, members of which are defined as aerobic, spore-forming and rod-shaped bacteria. Accepted 18 October 2018 However, some taxa are anaerobic, asporogenic and coccoid. 16S rRNA gene phylogeny is also unable to elucidate the taxonomic positions of several families incertae sedis within this order. Whole genome- Keywords: based phylogenetic approaches may provide a more accurate means to resolve higher taxonomic levels. A Bacillales Lactobacillales suite of phylogenomic approaches were applied to re-evaluate the taxonomy of 80 representative taxa of Bacillaceae eight families (and six family incertae sedis taxa) within the order Bacillales. -

Optimization and Immobilization of Amylase Obtained from Halotolerant Bacteria Isolated from Solar Salterns
Journal of Genetic Engineering and Biotechnology (2012) 10, 201–208 Academy of Scientific Research & Technology and National Research Center, Egypt Journal of Genetic Engineering and Biotechnology www.elsevier.com/locate/jgeb ARTICLE Optimization and immobilization of amylase obtained from halotolerant bacteria isolated from solar salterns Anbazhagan Mageswari a, Parthiban Subramanian b, Suganthi Chandrasekaran a, Karthikeyan Sivashanmugam a, S. Babu a, K.M. Gothandam a,* a School of Bio Sciences and Technology, VIT University, Vellore 632014, India b Department of Environmental and Biological Chemistry, Chungbuk National University, Cheongju, Chungbuk 361-763, Republic of Korea Received 11 August 2012; revised 14 September 2012; accepted 29 September 2012 Available online 2 November 2012 KEYWORDS Abstract The objective of the present study was to isolate halotolerant bacteria from the sediment Pontibacillus chungwhensis; sample collected from Marakanam Solar Salterns, Tamil Nadu, India using NaCl supplemented Bacillus barbaricus; media and screened for amylase production. Among the 22 isolates recovered, two strains that Solar saltern; had immense potential were selected for amylase production and designated as P1 and P2. The phy- Optimization; logenetic analysis revealed that P1 and P2 have highest homology with Pontibacillus chungwhensis Immobilization (99%) and Bacillus barbaricus (100%). Their amylase activity was optimized to obtain high yield under various temperature, pH and NaCl concentration. P1 and P2 strain showed respective, amy- lase activity maximum at 35 °C and 40 °C; pH 7.0 and 8.0; 1.5 M and 1.0 M NaCl concentration. Further under optimized conditions, the amylase activity of P1 strain (49.6 U mLÀ1) was higher than P2 strain. -

Ecology of Bacillaceae
Ecology of Bacillaceae INES MANDIC-MULEC,1 POLONCA STEFANIC,1 and JAN DIRK VAN ELSAS2 1University of Ljubljana, Biotechnical Faculty, Department of Food Science and Technology, Vecna pot 111, 1000 Ljubljana, Slovenia; 2Department of Microbial Ecology, Centre for Ecological and Evolutionary Studies, University of Groningen, Linneausborg, Nijenborgh 7, 9747AG Groningen, Netherlands ABSTRACT Members of the family Bacillaceae are among the been found in soil, sediment, and air, as well as in un- most robust bacteria on Earth, which is mainly due to their ability conventional environments such as clean rooms in the to form resistant endospores. This trait is believed to be the Kennedy Space Center, a vaccine-producing company, key factor determining the ecology of these bacteria. and even human blood (1–3). Moreover, members of However, they also perform fundamental roles in soil ecology (i.e., the cycling of organic matter) and in plant health and the Bacillaceae have been detected in freshwater and growth stimulation (e.g., via suppression of plant pathogens and marine ecosystems, in activated sludge, in human and phosphate solubilization). In this review, we describe the high animal systems, and in various foods (including fer- functional and genetic diversity that is found within the mented foods), but recently also in extreme environ- Bacillaceae (a family of low-G+C% Gram-positive spore-forming ments such as hot solid and liquid systems (compost bacteria), their roles in ecology and in applied sciences related and hot springs, respectively), salt lakes, and salterns (4– to agriculture. We then pose questions with respect to their 6). Thus, thermophilic genera of the family Bacillaceae ecological behavior, zooming in on the intricate social behavior that is becoming increasingly well characterized for some dominate the high-temperature stages of composting members of Bacillaceae.