Genetic and Morphological Analyses
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
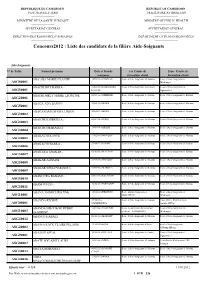
Concours2012 : Liste Des Candidats De La Filière Aide-Soignants
REPUBLIQUE DU CAMEROUN REPUBLIC OF CAMEROON PAIX-TRAVAIL-PATRIE PEACE-WORK-FATHERLAND ----------------------- ----------------------- MINISTERE DE LA SANTE PUBLIQUE MINISTRY OF PUBLIC HEALTH ----------------------- ----------------------- SECRETARIAT GENERAL SECRETARIAT GENERAL ----------------------- ----------------------- DIRECTION DES RESSOURCES HUMAINES DEPARTMENT OF HUMAN RESOURCES ----------------------- ----------------------- Concours2012 : Liste des candidats de la filière Aide-Soignants Aide-Soignants: N° de Table Nomset prénoms Date et lieu de 1er Centre de 2ème Centre de naissance formation choisi formation choisi ABA ABA MARIE CLAUDE 12/09/84 à ENONGAL Ecole d'Aide-Soignants d'Ebolowa Ecole d'Aide-Soignants de ASG80001 Mbalmayo ABACHONLE BARKA - 24/06/93 à KORO KORO Ecole d'Aide-Soignants de Garoua Ecole d'Aide-Soignants de ASG30001 KALFOU Ngaoundéré ABADIE MELY MARIE LEONTINE 08/03/88 à NDELELE Ecole d'Aide-Soignants de Garoua Ecole d'Aide-Soignants de Batouri ASG20001 ABAGA ADA BANYO 29/06/92 à MORA Ecole d'Aide-Soignants de Garoua Ecole d'Aide-Soignants de Maroua ASG50001 ABAGANAMA MAMA LIMAN 20/05/87 à MORA Ecole d'Aide-Soignants de Maroua Ecole d'Aide-Soignants de Garoua ASG30002 ABAÏCHO DJIBRILLA - 01/02/94 à BODO Ecole d'Aide-Soignants de Maroua Ecole d'Aide-Soignants de Garoua ASG30003 ABAICHO MAHAMAT - 13/09/90 à AFADE Ecole d'Aide-Soignants de Maroua Ecole d'Aide-Soignants de Garoua ASG30004 ABAKACHI KANTE 14/02/88 à WOULKY Ecole d'Aide-Soignants de Maroua Ecole d'Aide-Soignants de Garoua ASG30005 ABAKACHI BARKA -

Indian Native Games
Initiative for Moral and Cultural Training Foundation (IMCTF) Indian Native Games Indian Native Games 1 Details Book Name : Indian Native Games Edition : 2015 Pages : 280 Size : Demmy 1/8 Published by : Initiative for Moral and Cultural Training Foundation (IMCTF) Head Office : 4th Floor, Ganesh Towers, 152, Luz Church Road, Mylapore, Chennai - 600 004. Admin Office : 2nd Floor, “Gargi”, New No.6, (Old No.20) Balaiah Avenue, Luz, Mylapore, Chennai - 600 004. Email : [email protected], This book is available on Website : www.imct.org.in Printed by : Shri Vignesh Prints Chennai - 83 © Copy Rights to IMCTF 2 Indian Native Games CONTENT Sl. Topic Pg No. No. Class: I 1. Madurai Board - 17 2. Paramapadham - 19 3. Aakku paakku vethala paakku - 21 4. Athali Puthali - 22 5. Kaaya Pazhama - 23 6. Kokku para para - 24 7. Paruppu Kadayal - 25 8. Thoppukaranam - 26 9. Voothal - 27 10. Yoga 1) Pawanmuktasana - 27 2) Hasta utthanasana - 29 3) Sukha pranayama - 30 11. Killa parandi - 31 12. Kulaiyakulaiya mundirikkai - 32 13. Nadai Vandi Ottam - 33 14. Potato race I - 34 15. Train game - 35 Class: II 1. Sonaikattam - 37 2. Vatta Pallanguzhi(Brainvita) - 39 3. Achu Pootu - 40 4. Ainthu Pandhu - 41 5. Color color what color - 42 6. Concentration game - 43 7. Kannu moodi porul solle - 43 8. Kombu varaithal - 44 9. Nellikkai Vilaiyattu ( Isai nalkkali) - 45 10. Yoga 1) Pawanmuktasana - 46 2) Akarna Dhanurasana - 49 3) Dirgha Pranayama - 51 11. Kalla manna - 51 Indian Native Games 3 Sl. Topic Pg No. No. 12. Kappal viduthal - 52 13. Kichu Kichu thambalam - 53 14. Kulam kara - 55 15. -

2017 MAJOR EURO Music Festival CALENDAR Sziget Festival / MTI Via AP Balazs Mohai
2017 MAJOR EURO Music Festival CALENDAR Sziget Festival / MTI via AP Balazs Mohai Sziget Festival March 26-April 2 Horizon Festival Arinsal, Andorra Web www.horizonfestival.net Artists Floating Points, Motor City Drum Ensemble, Ben UFO, Oneman, Kink, Mala, AJ Tracey, Midland, Craig Charles, Romare, Mumdance, Yussef Kamaal, OM Unit, Riot Jazz, Icicle, Jasper James, Josey Rebelle, Dan Shake, Avalon Emerson, Rockwell, Channel One, Hybrid Minds, Jam Baxter, Technimatic, Cooly G, Courtesy, Eva Lazarus, Marc Pinol, DJ Fra, Guim Lebowski, Scott Garcia, OR:LA, EL-B, Moony, Wayward, Nick Nikolov, Jamie Rodigan, Bahia Haze, Emerald, Sammy B-Side, Etch, Visionobi, Kristy Harper, Joe Raygun, Itoa, Paul Roca, Sekev, Egres, Ghostchant, Boyson, Hampton, Jess Farley, G-Ha, Pixel82, Night Swimmers, Forbes, Charline, Scar Duggy, Mold Me With Joy, Eric Small, Christer Anderson, Carina Helen, Exswitch, Seamus, Bulu, Ikarus, Rodri Pan, Frnch, DB, Bigman Japan, Crawford, Dephex, 1Thirty, Denzel, Sticky Bandit, Kinno, Tenbagg, My Mate From College, Mr Miyagi, SLB Solden, Austria June 9-July 10 DJ Snare, Ambiont, DLR, Doc Scott, Bailey, Doree, Shifty, Dorian, Skore, March 27-April 2 Web www.electric-mountain-festival.com Jazz Fest Vienna Dossa & Locuzzed, Eksman, Emperor, Artists Nervo, Quintino, Michael Feiner, Full Metal Mountain EMX, Elize, Ernestor, Wastenoize, Etherwood, Askery, Rudy & Shany, AfroJack, Bassjackers, Vienna, Austria Hemagor, Austria F4TR4XX, Rapture,Fava, Fred V & Grafix, Ostblockschlampen, Rafitez Web www.jazzfest.wien Frederic Robinson, -

Payments for Environmental Services: Background
CHAPTER 8 A NEW TOOL FO R SUS ta IN A BLE FO R ES T MA N A GEMEN T IN CEN tra L AF R I ca : PA YMEN T S FO R ENVI R ONMEN ta L SE R VI C ES Guillaume Lescuyer, Alain Karsenty and Richard Eba’a Atyi Payments for Environmental Services: Background Tropical forests represent one of the rare eco- systems able to provide an abundance of products and support a diversity of human practices: from villagers seeking a source of natural products, the state seeking to conserve biodiversity, the timber processor, to the Global Environment Facility, which sees it as a carbon sink, the rainforest is multi-purpose and multi-stakeholder par excel- lence. First and foremost, the tropical forest provides material support for local populations’ way of life: the ecosystem is both their environment, a source of raw materials and foodstuffs, and a land reserve Ribas - GTZ © Frank for farming expansion. Most people in the Congo Photo 8.1: Tropical forests Basin meet basic needs by direct exploitation of alternate between dense their environment: fuelwood, timber, game, non- undergrowth and gaps. timber forest products (NTFP) … Nationally, the rainforest is often viewed as Table 8.1: Categories of environmental services provided by forests supporting economic development: industrial logging is supposed to generate economic growth, Regulatory functions Productive functions employment and foreign exchange earnings. The forest provides support to economic The forest provides basic resources, Most reforms of sub-regional forest policy in the activities and human well-being by: notably: past fifteen years were primarily geared towards - climate regulation - building materials: wood, lianas.. -

Exploring Farmers' Vulnerabilitiy and Agrobiodiversity in Perspective Of
Forbi Preasious Funwi et al. Exploring farmers’ vulnerability and agrobiodiversity in perspective of adaptation in Southern Cameroon Forbi Preasious Funwiab, Denis Jean Sonwac, William Armand Malab, Takanori Oishid, Marlene Toukam Ngansopab, and Marie Mbolob a Millennium Ecologic Museum, Cameroon b Department of Plant Biology, University of Yaounde I, Cameroon c Center for International Forestry Research (CIFOR), Central African Regional Office, Cameroon d African Studies Center – Tokyo University of Foreign Studies, Japan Abstract Climate change is a global phenomenon that indiscriminately affects all sectors of the economy and social life-support systems. New trends in climate change will leave high impacts on rural populations, whose livelihoods depend on agriculture and natural resources, leaving them increasingly vulnerable. Agrobiodiversity management is a promising method of facilitating adaptation to climatic changes. Hence, this study aimed to investigate the vulnerability of farmers and assess agrobiodiversity in Southern Cameroon in the context of adaptation. Focus groups and surveys were conducted in 31 villages in Ayos and Bokito in Southern Cameroon. The vulnerability index was computed for selected indicators of different components of vulnerability (exposure, sensitivity, and adaptive capacity). Data analysis revealed that in the two communities, the majority of villages were moderately vulnerable to climate change. However, Bokito community appeared to be more vulnerable than Ayos community. Farmers adopted several climate adaptation strategies such as crop replacement, replanting, planting of trees, cultivation of crops in swampy areas, and the expansion of cocoa cultivation in savannahs. Rich agrobiodiversity was identified in both sites; however, Ayos was richer than Bokito for wild plants, wildlife, and fisheries resources. The Bokito community also had a higher dependence on agriculture. -
Town and Countryside in a Dutch Perspective
Town and countryside in a Dutch perspective Town and countryside in a Dutch perspective Pim Kooij Nederlands Agronomisch Historisch Instituut Groningen/Wageningen 2010 Historia Agriculturae 42 Published by: Nederlands Agronomisch Historisch Instituut (NAHI) Oude Kijk in ’t Jatstraat 26, 9712 EK Groningen Hollandseweg 1, 6706 KN Wageningen Internet: www.rug.nl/let/nahi E-mail: [email protected] Photo cover: Studio Gaaikema, Groningen. Picture cover: Grote Markt Groningen, 1810 by H.L. Mijling (Groninger Museum). ISBN 978-90-367-4143-9 © 2010 Nederlands Agronomisch Historisch Instituut All rights reserved. No part of this book may be stored in a computerized system or reproduced in any form, by print, photo print, microfilm or any other means without written permission from the publisher. Cover design: Frank de Wit Print: Grafisch Centrum Rijksuniversiteit Groningen Production: Erwin H. Karel Contents Preface 1 1 Urbanization. What’s in a name? 5 2 Peripheral cities and their regions in the Dutch urban system until 29 1900 3 Urban history in the Netherlands 45 4 The images of Dutch cities in the nineteenth and twentieth 51 centuries 5 Where the action is. The introduction and acceptance of 67 infrastructural innovations in Dutch cities 1850-1950 6 Migrants in Dutch cities at the end of the nineteenth century 81 7 The destruction of Dutch cities during the Second World war 101 8 The Randstad conurbation: a floating metropolis in the Dutch 111 Delta (co-author Paul van der Laar) 9 Groningen: central place and peripheral city 135 10 Fingerprints of an urban elite: the case of a Dutch city in the 163 nineteenth century 11 Dutch and Russian regions compared. -
HIP-HOP FEST FLAP ‘Park’ Rapper’S Delight Barred for Bard Clyde by Ariella Cohen Stage a Polish Rendition of That Scottish Play
ANTI-YARDS SUIT LOSING STEAM? P.4 Brooklyn’s Real Newspaper BrooklynPaper.com • (718) 834–9350 • Brooklyn, NY • ©2007 BROOKLYN HEIGHTS–DOWNTOWN EDITION AWP/16 pages • Vol. 30, No. 38 • Saturday, Sept. 29, 2007 • FREE INCLUDING DUMBO HIP-HOP FEST FLAP ‘Park’ rapper’s delight barred for Bard Clyde By Ariella Cohen stage a Polish rendition of that Scottish play. opment will finance greenspace along a 1.3-mile The Brooklyn Paper Festival organizers believe the move was racial- stretch from DUMBO to the foot of Atlantic Av- ly motivated. enue. Opponents believe that public events will not A production of Shakespeare’s “Macbeth” will “Hip hop brings a lot more brown people to this be public at all, but subject to the whims of the replace a hip-hop festival next summer in a DUM- neighborhood, and people who live here are not wealthy condo-dwellers whose maintenance fees BO venue controlled by the Brooklyn Bridge Park comfortable with it,” said Wes Jackson, whose will pay for the park’s upkeep. Conservancy — and organizers of the rap show Room Service Production founded the festival in “What we have feared all along is that the Conser- believe that race played a role. 2005. vancy, which has received enormous amounts of fi- The Brooklyn Hip-Hop Festival — which “[People have told me that residents say], ‘The nancial support from Brooklyn Heights residents, will brought thousands of people and big-name rappers festival should be in Commodore Barry Park be- run the park for their benefit and not the benefit of res- to the park-and-condo waterfront development site tween the projects and the BQE, not next to my idents of other neighborhoods,” said Roy Sloane, a Inky in 2006 and 2007— had already scheduled its 2008 $2.5-million condo.’ ” former president of the Cobble Hill Association. -

Violence Against Women in PNG?
TABLE OF CONTENTS 1. Introduction ............................................................................................................. 1 2. Background .............................................................................................................. 3 3. What is Known About Violence Against Women in PNG? ................................. 5 3.1 Available Data in Papua New Guinea ............................................................. 5 3.2 Steps to Improve Data Collection................................................................... 8 3.3 Why Data Collection Matters ........................................................................... 9 4. Where and How do Women Experience Violence? .......................................... 10 4.1 International Definitions of Violence Against Women ............................... 11 4.2 Domestic Violence ........................................................................................... 12 Intimate Partner Violence ............................................................................. 12 Sexual Violence in Intimate Relationships ................................................. 13 Women as Perpetrators – Violence Against Husbands? .......................... 14 Women as Perpetrators - Violence Between Co-Wives ........................... 15 4.3 Sexual Violence ............................................................................................... 15 Rape and Gang Rape ................................................................................... -

ARCRP2014.Pdf
CONTENTS Abbreviations ...................................................................................................................................................... 2 A. KEY MESSAGES .................................................................................................................................. 4 B. IMPACT PATHWAY AND INTERMEDIATE DEVELOPMENT OUTCOMES ................................ 5 C. PROGRESS ALONG THE IMPACT PATHWAY ................................................................................ 6 D. GENDER RESEARCH ACHIEVEMENTS ......................................................................................... 11 E. PARTNERSHIP BUILDING ACHIEVEMENTS ................................................................................ 12 F. CAPACITY BUILDING ...................................................................................................................... 13 G. RISK MANAGEMENT........................................................................................................................ 13 H. LESSONS LEARNED ......................................................................................................................... 14 I. CRP FINANCIAL REPORT ................................................................................................................ 14 Annex 1: Indicator Table………………………………………………………………………………………15 Annex 2: Technical Progress Report 2014…………………………………………………………………….27 Annex 3: Publications in 2014………………………………………………………………………………..139 Annex 4: Gender -

CGIAR Research Program on Forests, Trees and Agroforestry
CONTENTS Abbreviations ...................................................................................................................................................... 2 A. KEY MESSAGES .................................................................................................................................. 4 B. IMPACT PATHWAY AND INTERMEDIATE DEVELOPMENT OUTCOMES ................................ 5 C. PROGRESS ALONG THE IMPACT PATHWAY ................................................................................ 6 D. GENDER RESEARCH ACHIEVEMENTS ......................................................................................... 11 E. PARTNERSHIP BUILDING ACHIEVEMENTS ................................................................................ 12 F. CAPACITY BUILDING ...................................................................................................................... 13 G. RISK MANAGEMENT........................................................................................................................ 13 H. LESSONS LEARNED ......................................................................................................................... 14 I. CRP FINANCIAL REPORT ................................................................................................................ 14 Annex 1: Indicator Table………………………………………………………………………………………15 Annex 2: Technical Progress Report 2014…………………………………………………………………….27 Annex 3: Publications in 2014………………………………………………………………………………..139 Annex 4: Gender -

Lakeland, FL April 6-11Th 2021
Lakeland, FL April 6-11th 2021 Elite 8 Petite Dancer Avery Buck - Rose's Turn - Central Florida Dance Center 1st Runner-Up - Khloe Brooks - Salute - Central Florida Dance Center 2nd Runner-Up - Mackenzie Cromwell - Me And The Sky - The Dance Collective Elite 8 Junior Dancer Sara Boutwell - Sparrow - The Dance Collective 1st Runner-Up - Brooke Zatko - What About Us - BSDA 2nd Runner-Up - Hannah Harris - Pixie Dust - Central Florida Dance Center Elite 8 Teen Dancer Ava Johnson - Ain't No Sunshine - Central Florida Dance Center 1st Runner-Up - Gisselle Quinones - Feeling Good - BSDA 2nd Runner-Up - Vadri Olivella-Romero - Bohemian Rhapsody - Elite Dance Center Elite 8 Senior Dancer Tori Wegner - Babette - BSDA 1st Runner-Up - Chloe Finch - We - East Coast Performing Arts 2nd Runner-Up - Elle Skoglund - Trying My Best - A&G Dance Academy Top Select Petite Solo 1 - Abigail Lee - I Fell - BSDA 2 - Natalie Kim - Save Your Life - West Florida Dance Company 3 - Alyssa Pinzon - Good Things - Victorias School of Dance 4 - Avery Buck - Rose's Turn - Central Florida Dance Center 5 - Mackenzie Cromwell - Me And The Sky - The Dance Collective Top Select Junior Solo 1 - Isabella Bustillo - Shake Senora - The Dance Collective 2 - Sara Boutwell - Sparrow - The Dance Collective 3 - Alyessia Chiavatti - I Will Wait - New Level Dance Company 4 - Bailey Roncevich - Greed - Central Florida Dance Center 5 - Bailey Toney - Imagine - Dance Unlimited of Highlands County 6 - Hannah Harris - Pixie Dust - Central Florida Dance Center 7 - Presley Loftis - Daughter -

Journal of Berggorilla & Regenwald Direkthilfe
Gorilla Journal Journal of Berggorilla & Regenwald Direkthilfe No. 54, June 2017 A Population Survey of the One of the Biggest A Tribute to Colo Estimate of Great Cross River Ape Traffickers of Apes in Itombwe Gorilla at Tofala Africa Arrested Hill BERGGORILLA & REGENWALD DIREKTHILFE Authors of this Issue returned to Cameroon to collect data CONTENTS for his doctoral thesis on the Cross D. R. Congo 3 Gedeon Banswe is the GIS and data River gorillas. Conservation and Sensitization base officer with the WWF Itombwe Dr. Miki Matsubara took part in the Activities at Sarambwe 3 Program. gorilla census of Kahuzi-Biega in 1996 A Population Estimate of Great Andrew Dunn is Project Manager for and of Petit Loango in Gabon in 1998. Apes in Itombwe 5 the WCS biodiversity research program She studied social relationships of Community Development Projects in southeastern Nigeria. He has been juvenile gorillas in the Howletts and near Mount Tshiaberimu 13 working on biological survey and con- Port Lympne Wild Animal Parks. Now Community Conservation for the servation projects in Africa since 1989. she is an adjunct lecturer of Chukyo Park and Local Communities 14 Prof. Colin P. Groves wrote his University. Rwanda 17 Ph.D. thesis on gorilla osteology and Menard Mbende is the WWF DRC Cantsbee – a Gorilla Legend 17 taxonomy. After working at American protected areas Program Manager. He Death of Three Silverbacks 17 and British universities, he emigrated has been involved in the 2015 Itombwe Cross River 18 to Australia in 1974. Now he is retired, survey. Update on the Proposed after teaching primatology and human Dr.