2019 Summer Undergraduate Research Program
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Late Calcium EDTA Rescues Hippocampal CA1 Neurons from Global Ischemia-Induced Death
The Journal of Neuroscience, November 3, 2004 • 24(44):9903–9913 • 9903 Neurobiology of Disease Late Calcium EDTA Rescues Hippocampal CA1 Neurons from Global Ischemia-Induced Death Agata Calderone, Teresa Jover,* Toshihiro Mashiko,* Kyung-min Noh, Hidenobu Tanaka,† Michael V. L. Bennett, and R. Suzanne Zukin Department of Neuroscience, Albert Einstein College of Medicine, Bronx, New York 10461 Transient global ischemia induces a delayed rise in intracellular Zn 2ϩ, which may be mediated via glutamate receptor 2 (GluR2)-lacking AMPA receptors (AMPARs), and selective, delayed death of hippocampal CA1 neurons. The molecular mechanisms underlying Zn 2ϩ toxicity in vivo are not well delineated. Here we show the striking finding that intraventricular injection of the high-affinity Zn 2ϩ chelator calcium EDTA (CaEDTA) at 30 min before ischemia (early CaEDTA) or at 48–60 hr (late CaEDTA), but not 3–6 hr, after ischemia, afforded robust protection of CA1 neurons in ϳ50% (late CaEDTA) to 75% (early CaEDTA) of animals. We also show that Zn 2ϩ acts via temporally distinct mechanisms to promote neuronal death. Early CaEDTA attenuated ischemia-induced GluR2 mRNA and protein downregulation (and, by inference, formation of Zn 2ϩ-permeable AMPARs), the delayed rise in Zn 2ϩ, and neuronal death. These findings suggest that Zn 2ϩ acts at step(s) upstream from GluR2 gene downregulation and implicate Zn 2ϩ in transcriptional regulation and/or GluR2 mRNA stability. Early CaEDTA also blocked mitochondrial release of cytochrome c and Smac/DIABLO (second mitochondria-derived activator of caspases/direct inhibitor of apoptosis protein-binding protein with low pI), caspase-3 activity (but not procaspase-3 cleavage), p75 NTR induction, and DNA fragmentation. -

Department of Neuroscience Faculty Research Interests at the Albert Einstein
Dominick P. Purpura Department of Neuroscience Faculty Research Interests at the Albert Einstein DOBRENIS EMMONS ESKANDER COEN-CAGLI College of Medicine FISHMAN CHUA FRANCESCONI CASTILLO 2019–2020 FRENETTE BUSCHKE FRICKER GALANOPOULOU BUELOW GONÇALVES BERGMAN HALL BENNETT HÉBERT BATISTA-BRITO JORDAN BALLABH KHODAKHAH AUTRY ASCHNER AREZZO KURSHAN ALPERT AKABAS LACHMAN LAROCCA LEGATT LIPTON MEHLER MOLHOLM MOSHÉ NICOLA PEÑA PEREDA ROSS RUDOLPH SCHWARTZ SECOMBE SHARP SINGER SJULSON SOLDNER SPRAY STEINSCHNEIDER SUADICANI SUSSMAN VERSELIS WALKLEY WILSON YANG ZHENG ZUKIN Dominick P. Purpura Department of Neuroscience Faculty Research Interests at the Albert Einstein College of Medicine 2019–2020 Myles Akabas, M.D., Ph.D. 1 Peri Kurshan, Ph.D. 68 Joseph C. Arezzo, Ph.D. 6 Herbert M. Lachman, M.D. 70 Michael Aschner, Ph.D. 8 Jorge N. Larocca, Ph.D. 73 Anita E. Autry, Ph.D. 10 Alan D. Legatt, M.D., Ph.D. 75 Praveen Ballabh, M.D. 12 Michael L. Lipton, M.D., Ph.D. 77 Renata Batista-Brito, Ph.D. 14 Mark F. Mehler, M.D. 81 Michael V. L. Bennett, D.Phil. 16 Sophie Molholm, Ph.D. 83 Aviv Bergman, Ph.D. 18 Solomon L. Moshé, M.D. 89 Herman Buschke, M.D. 24 Saleem M. Nicola, Ph.D. 93 Pablo E. Castillo, M.D., Ph.D. 25 José L. Peña, M.D., Ph.D. 96 Streamson C. Chua, Jr., M.D., Ph.D. 27 Alberto E. Pereda, M.D., Ph.D. 98 Ruben Coen-Cagli, Ph.D. 29 Rachel A. Ross, M.D., Ph.D. 100 Kostantin Dobrenis, Ph.D. 31 Stephanie Rudolph, Ph.D. 102 Scott W. -

TLN Agenda-101507
Fall 2007 Janelia Conference: Translation at the Synapse Full Schedule Sunday, October 21st 3:00 pm Check-in 6:00 pm Reception 7:00 pm Dinner 8:00 pm Session 1 (chair - Kevin Moses) 8:00 pm Oswald Steward, University of California, Irvine Mechanisms underlying the selective localization of Arc mRNA at active synapses 8:30 pm James Eberwine, University of Pennsylvania School of Medicine Molecular Biology of the Neuronal Dendrite 9:00 pm Robert Singer, Albert Einstein College of Medicine Following single mRNAs from birth to translation 9:30 pm Erin Schuman, California Institute of Technology/HHMI Visualization of dendritic protein synthesis 10:00 pm Refreshments available at Bob’s pub Fall 2007 Janelia Conference: Translation at the Synapse Monday, October 22nd 7:30 am Breakfast 8:30 am Session 2 (chair - Erin Schuman) 8:30 am Michael Kiebler, Medical University of Vienna The role of Staufen in dendritic RNP transport and dendritic spine morphogenesis 9:00 am Nobutaka Hirokawa, University of Tokyo Intracellular transport of mRNA and kinesin superfamily proteins, KIFs 9:30 am Mark F. Bear, Massachusetts Institute of Technology/HHMI Fragile X syndrome: A disease of synaptic protein synthesis 10:00 am Suzanne Zukin, Albert Einstein College of Medicine AMPA receptor mRNA trafficking in dendrites and synaptic plasticity: dysregulation in Fragile X 10:30 am Break and Group Photo 11:00 am Gary J. Bassell, Emory University School of Medicine Dysregulated mGluR-dependent translation of AMPA receptor and PSD- 95 mRNAs in fragile x syndrome 11:30 am Tom Jongens, University of Pennsylvania School of Medicine Regulation of the Drosophila Fragile X mental retardation gene by the siRNA pathway 12:00 pm Claudia Bagni, University of Rome Translational control at synapses and mental retardation: new insights into the Fragile X Syndrome 12:30 pm Lunch 1:00 pm Tour 2:00 pm Session 3 (chair - Mark F. -

WCBR Program3
Welcome to the Thirty-Fifth Annual Winter Conference on Brain Research The Winter Conference on Brain Research (WCBR) was founded in 1968 to promote free exchange of information and ideas within neuroscience. It was the intent of the founders that both formal and informal interactions would occur between clinical and laboratory based neuroscientists. During the past thirty years neuroscience has grown and expanded to include many new fields and methodologies. This diversity is also reflected by WCBR participants and in our program. A primary goal of the WCBR is to enable participants to learn about the current status of areas of neuroscience other than their own. Another objective is to provide a vehicle for scientists with common interests to discuss current issues in an informal setting. On the other hand, WCBR is not designed for presentations limited to communicating the latest data to a small group of specialists; this is best done at national society meetings. The program includes panels (reviews for an audience not neces- sarily familiar with the area presented), workshops (informal discussions of current issues and data), and a number of posters. The annual conference lecture will be presented at the Sunday breakfast on Sunday, January 27. Our guest speaker will be Dr. Donald Kennedy, Editor-in-Chief of Science. On Tuesday, January 29, a town meeting will be held for the Aspen/Snowmass commu- nity at which Dr. George Ricaurte, and WCBR participants will discuss drug addiction and toxicity of addictive drugs. Also, participants in the WCBR Outreach Program will present sessions at local schools throughout the week to pique students’ interest in science. -

WCBR Program 04
Welcome to the Thirty-Seventh Annual Winter Conference on Brain Research The Winter Conference on Brain Research (WCBR) was founded in 1968 to promote free exchange of information and ideas within neuroscience. It was the intent of the founders that both formal and informal interactions would occur between clinical and laboratory-based neuroscientists. During the past thirty years neuroscience has grown and expanded to include many new fields and methodologies. This diversity is also reflected by WCBR participants and in our program. A primary goal of the WCBR is to enable participants to learn about the current status of areas of neuroscience other than their own. Another objective is to provide a vehicle for scientists with common interests to discuss current issues in an informal setting. On the other hand, WCBR is not designed for presentations limited to communicating the latest data to a small group of specialists; this is best done at national society meetings. The program includes panels (reviews for an audience not neces- sarily familiar with the area presented), workshops (informal discussions of current issues and data), and a number of posters. The annual conference lecture will be presented at the Sunday breakfast on January 25. Our guest speaker will be The Honorable John Edward Porter, former Congressman from Illinois and Chair of the House Appropriations Committee. The title of his talk will be What’s Going on in Washington: We Need to Talk! On Tuesday, January 27, a town meeting will be held for the Copper Mountain community, at which Dr. Michael Zigmond, Director of the Morris K. -

2018 Abstract Book
Graduate Division of Biomedical Sciences SUMMER UNDERGRADUATE RESEARCH PROGRAM 2018 Victoria H. Freedman, Ph.D. Associate Dean for Graduate Programs Director, Summer Undergraduate Research Program 2018 Summer Undergraduate Research Program Student Name Undergraduate School Mentor Name Abdurakhmon Abdulatipov University of North Carolina-Charlotte Chandan Guha Rahma Ahmed Bard College Andras Fiser Saad Ahmed CUNY-City College Steven Almo Olivia Albert New York Institute of Technology David Spray Michael Angelov CUNY-Hunter College Yair Botbol Mone Anzai Emory University Anna Francesconi Elana Apfelbaum Yeshiva University Libusha Kelly Alyssa Azuma Arizona State University Britta Will Kayla Baker Tuskegee University Felipe Diaz-Griffero Corin Bell Spelman College David Sharpe Sasha Bonilla CUNY-Brooklyn College R. Suzanne Zukin Leslieann Diaz CUNY-Lehman College Jon Lai Sarah Duggan Amherst College Derek Huffman Michel Fallah CUNY-Brooklyn College Anita Autry-Dixon Cara Ford Xavier University of Louisiana Kimberly Reidy Nathan Frederick Bates College Sridhar Mani Antonio Freitas CUNY-Hunter College Saleem Nicola Lauren Fries Oberlin College Hannes Buelow Bailey Frolich Yeshiva University Michael Ashner Jiselle Gill St. John's University Nicholas Baker Lauren Goodes Xavier University of Louisiana Sophia Molholm Jennifer Guarino University Of Chicago Bridgit Shafit-Zagardo Tanjanay Hardy Spelman College John Greally Christopher Hiner University Of Maryland-College Park Jonathan Baker Jayme Jackson University Of Arizona Margaret Kielian Fred -

A Silent Synapse-Based Mechanism for Cocaine-Induced Locomotor Sensitization
The Journal of Neuroscience, June 1, 2011 • 31(22):8163–8174 • 8163 Cellular/Molecular A Silent Synapse-Based Mechanism for Cocaine-Induced Locomotor Sensitization Travis E. Brown,1* Brian R. Lee,1* Ping Mu,1 Deveroux Ferguson,2 David Dietz,2 Yoshinori N. Ohnishi,2 Ying Lin,3 Anna Suska,4,5 Masago Ishikawa,1 Yanhua H. Huang,1 Haowei Shen,6 Peter W. Kalivas,6 Barbara A. Sorg,1 R. Suzanne Zukin,3 Eric J. Nestler,2 Yan Dong,1 and Oliver M. Schlu¨ter4,5 1Program in Neuroscience, Washington State University, Pullman, Washington 99164-6520, 2Fishberg Department of Neuroscience, Mount Sinai School of Medicine, New York, New York 10029, 3Dominick P. Purpura Department of Neuroscience, Albert Einstein College of Medicine, New York NY 10460, 4Molecular Neurobiology, European Neuroscience Institute, 37077 Go¨ttingen, Germany, 5Molecular Physiology of the Brain, Go¨ttingen Graduate School for Neurosciences and Molecular Biosciences, 37077 Go¨ttingen, Germany, 6Department of Neuroscience, Medical University of South Carolina, Charleston, South Carolina 29425 Locomotor sensitization is a common and robust behavioral alteration in rodents whereby following exposure to abused drugs such as cocaine, the animal becomes significantly more hyperactive in response to an acute drug challenge. Here, we further analyzed the role of cocaine-induced silent synapses in the nucleus accumbens (NAc) shell and their contribution to the development of locomotor sensiti- zation. Using a combination of viral vector-mediated genetic manipulations, biochemistry, and electrophysiology in a locomotor sensi- tization paradigm with repeated, daily, noncontingent cocaine (15 mg/kg) injections, we show that dominant-negative cAMP-element binding protein (CREB) prevents cocaine-induced generation of silent synapses of young (30 d old) rats, whereas constitutively active CREB is sufficient to increase the number of NR2B-containing NMDA receptors (NMDARs) at synapses and to generate silent synapses. -

REST)-Dependent Epigenetic Remodeling Is Critical to Ischemia-Induced Neuronal Death
Repressor element-1 silencing transcription factor (REST)-dependent epigenetic remodeling is critical to ischemia-induced neuronal death Kyung-Min Noha,1, Jee-Yeon Hwanga,1, Antonia Follenzib, Rodoniki Athanasiadouc, Takahiro Miyawakia, John M. Greallyc,d, Michael V. L. Bennetta,2, and R. Suzanne Zukina,2 aDominick P. Purpura Department of Neuroscience, bDepartment of Pathology, cDepartment of Genetics, and dDepartment of Medicine, Albert Einstein College of Medicine, New York, NY 10461 Contributed by Michael V. L. Bennett, January 15, 2012 (sent for review December 22, 2011) Dysregulation of the transcriptional repressor element-1 silencing platforms that recruit histone deacetylases (HDACs) 1 and 2. transcription factor (REST)/neuron-restrictive silencer factor is HDACs deacetylate core histone proteins (16, 22). In addition, important in a broad range of diseases, including cancer, di- REST recruits the site-specific histone methyl-transferase G9a, abetes, and heart disease. The role of REST-dependent epigenetic which promotes dimethylation of histone 3 at lysine 9 (H3K9me2) modifications in neurodegeneration is less clear. Here, we show via CoREST-dependent (8) and independent (23) mechanisms; that neuronal insults trigger activation of REST and CoREST in the site-specific histone demethylase LSD1, which removes a clinically relevant model of ischemic stroke and that REST binds methyl groups from histone 3 mono- or dimethylated at lysine 4 a subset of “transcriptionally responsive” genes (gria2, grin1, (H3K4me1, HSK4me2) (24, 25); and methyl CpG binding protein chrnb2, nefh, nfκb2, trpv1, chrm4,andsyt6), of which the AMPA 2 (MeCP2) (8, 26), a protein that reads epigenetic marks on core receptor subunit GluA2 is a top hit. -

Bidirectional Control of Mrna Translation and Synaptic Plasticity
Bidirectional Control of mRNA Translation and Synaptic Plasticity by the Cytoplasmic Polyadenylation Complex Tsuyoshi Udagawa, University of Massachusetts Sharon Swanger, Emory University Koichi Takeuchi, Albert Einstein College of Medicine Jong Heon Kim, University of Massachusetts Vijayalaxmi Nalavadi, Emory University Jihae Shin, University of Massachusetts Lori J. Lorenz, University of Massachusetts R. Suzanne Zukin, Albert Einstein College of Medicine Gary Bassell, Emory University Joel D. Richter, University of Massachusetts Journal Title: Molecular Cell Volume: Volume 47, Number 2 Publisher: Elsevier (Cell Press): 12 month embargo | 2012-07-27, Pages 253-266 Type of Work: Article | Post-print: After Peer Review Publisher DOI: 10.1016/j.molcel.2012.05.016 Permanent URL: https://pid.emory.edu/ark:/25593/s9ft2 Final published version: http://dx.doi.org/10.1016/j.molcel.2012.05.016 Copyright information: © 2012 Elsevier Inc. This is an Open Access work distributed under the terms of the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (http://creativecommons.org/licenses/by-nc-nd/4.0/). Accessed September 25, 2021 11:20 AM EDT NIH Public Access Author Manuscript Mol Cell. Author manuscript; available in PMC 2013 July 27. NIH-PA Author ManuscriptPublished NIH-PA Author Manuscript in final edited NIH-PA Author Manuscript form as: Mol Cell. 2012 July 27; 47(2): 253–266. doi:10.1016/j.molcel.2012.05.016. Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex Tsuyoshi Udagawa1,*, Sharon A. Swanger2,*, Koichi Takeuchi3, Jong Heon Kim1,#, Vijayalaxmi Nalavadi2, Jihae Shin1, Lori J. Lorenz1, R. Suzanne Zukin3, Gary J. Bassell2,4, and Joel D. -
Activation of Autophagy Rescues Synaptic and Cognitive Deficits in Fragile X Mice
Activation of autophagy rescues synaptic and cognitive deficits in fragile X mice Jingqi Yana,1, Morgan W. Porcha,1, Brenda Court-Vazqueza, Michael V. L. Bennetta,2, and R. Suzanne Zukina,2 aDominick P. Purpura Department of Neuroscience, Albert Einstein College of Medicine, New York, NY 10461 Contributed by Michael V. L. Bennett, August 17, 2018 (sent for review May 29, 2018; reviewed by Claudia Bagni and Leonard K. Kaczmarek) Fragile X syndrome (FXS) is the most frequent form of heritable (17–19). Upon activation, mTOR phosphorylates Unc-51–like intellectual disability and autism. Fragile X (Fmr1-KO) mice exhibit autophagy-activating kinase 1 (ULK-1) at S757, a target of aberrant dendritic spine structure, synaptic plasticity, and cogni- mTORC1 and a well-established antiautophagy site (Fig. 1) tion. Autophagy is a catabolic process of programmed degradation (20). This, in turn, sequesters ULK-1 away from AMP kinase and recycling of proteins and cellular components via the lyso- (AMPK). AMPK phosphorylates and activates ULK-1 at S317. somal pathway. However, a role for autophagy in the pathophys- Upon activation, ULK-1 promotes phosphorylation and activa- iology of FXS is, as yet, unclear. Here we show that autophagic tion of Beclin-1 at S14, a critical step in the nucleation phase of flux, a functional readout of autophagy, and biochemical markers autophagy (21). Beclin-1 promotes lipidation of LC3-I to gen- of autophagy are down-regulated in hippocampal neurons of frag- erate its lipidated form LC3-II, enabling elongation of the lim- ile X mice. We further show that enhanced activity of mammalian iting membrane and the formation of autophagosomes (22). -
The Essential Role of AMPA Receptor Glua2 Subunit RNA Editing in the Normal and Diseased Brain
REVIEW ARTICLE published: 11 April 2012 MOLECULAR NEUROSCIENCE doi: 10.3389/fnmol.2012.00034 The essential role of AMPA receptor GluA2 subunit RNA editing in the normal and diseased brain Amanda Wright1,2 and Bryce Vissel1,2* 1 Neurodegenerative Disorders Laboratory, Neuroscience Department, Garvan Institute of Medical Research, Sydney, NSW, Australia 2 Faculty of Medicine, University of New South Wales, Sydney, NSW, Australia Edited by: α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors are comprised of R. Suzanne Zukin, Albert Einstein different combinations of GluA1–GluA4 (also known as GluR1–GluR4 and GluR-A to GluR-D) College of Medicine, USA subunits. The GluA2 subunit is subject to RNA editing by the ADAR2 enzyme, which con- Reviewed by: verts a codon for glutamine (Gln; Q), present in the GluA2 gene, to a codon for arginine Tim Green, University of 2+ Liverpool, UK (Arg; R) found in the mRNA. AMPA receptors are calcium (Ca )-permeable if they contain Lorna Role, SUNY Stony the unedited GluA2(Q) subunit or if they lack the GluA2 subunit. While most AMPA recep- Brook, USA tors in the brain contain the edited GluA2(R) subunit and are therefore Ca2+-impermeable, *Correspondence: recent evidence suggests that Ca2+-permeable AMPA receptors are important in synaptic Bryce Vissel, Neurodegenerative 2+ Disorders Laboratory, Neuroscience plasticity, learning, and disease. Strong evidence supports the notion that Ca -permeable Department, Garvan Institute of AMPA receptors are usually GluA2-lacking AMPA receptors, with little evidence to date Medical Research, 384 Victoria Street, for a significant role of unedited GluA2 in normal brain function. -
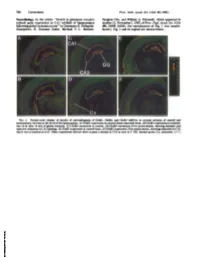
Neurobiology. in the Article "Switch in Glutamate Receptor Subunit
780 Corrections Proc. Natl. Acad. Sci. USA 90 (1993) Neurobiology. In the article "Switch in glutamate receptor Sunghee Cho, and William A. Pulsinelli, which appeared in subunit gene expression in CA1 subfield of hippocampus number 21, November 1, 1992, ofProc. Natl. Acad. Sci. USA following global ischemia in rats" by Domenico E. Pellegrini- (89, 10499-10503), the reproduction of Fig. 1 was unsatis- Giampietro, R. Suzanne Zukin, Michael V. L. Bennett, factory. Fig. 1 and its legend are shown below. FIG. 1. Pseudo-color display of density of autoradiograms of GluRi, GluR2, and GluR3 mRNAs in coronal sections of control and postischemic rat brain at the level ofthe hippocampus. (A) GluRl expression in control (sham-operated) brain. (B) GluRl expression in ischemic rats 24 hr after 10 min of global ischemia. (C) GluR2 expression in control. (D) GluR2 expression 24 hr postischemia, showing dramatic and selective reduction in CA1 labeling. (E) GluR3 expression in control brain. (F) GluR3 expression 24 hr postischemia, showing reduction in CA1 that is not as marked as in D. Other experiments did not show as great a decline in CA3 as seen in F. DG, dentate gyrus; Cx, neocortex. (x7.) Downloaded by guest on September 30, 2021 Proc. NatI. Acad. Sci. USA Vol. 89, pp. 10499-10503, November 1992 Neurobiology Switch in glutamate receptor subunit gene expression in CAl subfield of hippocampus following global ischemia in rats caina-at-mino-3-hydroxy-5-umethyl-4-lsoxazole bproplonic add/excdtotoixdty) DOMENICO E. PELLEGRINI-GIAMPIETRO*t, R. SUZANNE ZUKIN**, MICHAEL V. L. BENNETT*, SUNGHEE CHO§, AND WILLIAM A.