Stratified Analyses of Genome Wide Association Study
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
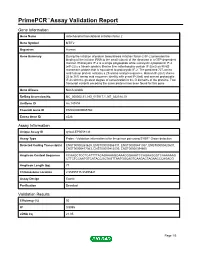
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name mitochondrial translational initiation factor 2 Gene Symbol MTIF2 Organism Human Gene Summary During the initiation of protein biosynthesis initiation factor-2 (IF-2) promotes the binding of the initiator tRNA to the small subunit of the ribosome in a GTP-dependent manner. Prokaryotic IF-2 is a single polypeptide while eukaryotic cytoplasmic IF-2 (eIF-2) is a trimeric protein. Bovine liver mitochondria contain IF-2(mt) an 85-kD monomeric protein that is equivalent to prokaryotic IF-2. The predicted 727-amino acid human protein contains a 29-amino acid presequence. Human IF-2(mt) shares 32 to 38% amino acid sequence identity with yeast IF-2(mt) and several prokaryotic IF-2s with the greatest degree of conservation in the G domains of the proteins. Two transcript variants encoding the same protein have been found for this gene. Gene Aliases Not Available RefSeq Accession No. NC_000002.11, NG_017017.1, NT_022184.15 UniGene ID Hs.149894 Ensembl Gene ID ENSG00000085760 Entrez Gene ID 4528 Assay Information Unique Assay ID qHsaCEP0058136 Assay Type Probe - Validation information is for the primer pair using SYBR® Green detection Detected Coding Transcript(s) ENST00000263629, ENST00000366137, ENST00000441307, ENST00000420637, ENST00000417363, ENST00000412530, ENST00000394600 Amplicon Context Sequence CCAAGCTCCTCATTTTACAGAAAAGGAAACGGAAATCCAGAAGGGTCAAAAAAG CTTCTCCAATGTCATACCGCTAGTTAATGGCAGTCAAGACTAGAACCCAGACG Amplicon Length (bp) 77 Chromosome Location 2:55495715-55495821 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 95 R2 0.9995 cDNA Cq 21.05 Page 1/5 PrimePCR™Assay Validation Report cDNA Tm (Celsius) 79 gDNA Cq 24.56 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

LIM Domain Proteins Pinch1/2 Regulate Chondrogenesis and Bone Mass in Mice
Bone Research www.nature.com/boneres ARTICLE OPEN LIM domain proteins Pinch1/2 regulate chondrogenesis and bone mass in mice Yiming Lei1, Xuekun Fu1, Pengyu Li1, Sixiong Lin1,2, Qinnan Yan1, Yumei Lai3, Xin Liu1, Yishu Wang1, Xiaochun Bai 4, Chuanju Liu5,6, Di Chen7, Xuenong Zou2, Xu Cao8, Huiling Cao1 and Guozhi Xiao1 The LIM domain-containing proteins Pinch1/2 regulate integrin activation and cell–extracellular matrix interaction and adhesion. Here, we report that deleting Pinch1 in limb mesenchymal stem cells (MSCs) and Pinch2 globally (double knockout; dKO) in mice causes severe chondrodysplasia, while single mutant mice do not display marked defects. Pinch deletion decreases chondrocyte proliferation, accelerates cell differentiation and disrupts column formation. Pinch loss drastically reduces Smad2/3 protein expression in proliferative zone (PZ) chondrocytes and increases Runx2 and Col10a1 expression in both PZ and hypertrophic zone (HZ) chondrocytes. Pinch loss increases sclerostin and Rankl expression in HZ chondrocytes, reduces bone formation, and increases bone resorption, leading to low bone mass. In vitro studies revealed that Pinch1 and Smad2/3 colocalize in the nuclei of chondrocytes. Through its C-terminal region, Pinch1 interacts with Smad2/3 proteins. Pinch loss increases Smad2/3 ubiquitination and degradation in primary bone marrow stromal cells (BMSCs). Pinch loss reduces TGF-β-induced Smad2/3 phosphorylation and nuclear localization in primary BMSCs. Interestingly, compared to those from single mutant mice, BMSCs from dKO mice express dramatically lower protein levels of β-catenin and Yap1/Taz and display reduced osteogenic but increased adipogenic differentiation capacity. Finally, ablating Pinch1 in chondrocytes and Pinch2 globally causes severe osteopenia with subtle limb fi 1234567890();,: shortening. -

Targeting of the EGFR/Β1 Integrin Connecting Proteins PINCH1 and Nck2 Radiosensitizes Three-Dimensional SCC Cell Cultures
ONCOLOGY REPORTS 34: 469-476, 2015 Targeting of the EGFR/β1 integrin connecting proteins PINCH1 and Nck2 radiosensitizes three-dimensional SCC cell cultures LYDIA ROSSOW1,2, IRIS EKE1,2, ELLEN DICKREuTER1,2 and NILS CORDES1-5 1OncoRay-National Center for Radiation Research in Oncology, Faculty of Medicine and university Hospital Carl Gustav Carus, Technische universität Dresden, D-01307 Dresden, and Helmholtz-Zentrum Dresden-Rossendorf, D-01328 Dresden; 2Department of Radiation Oncology, university Hospital Carl Gustav Carus, Technische universität Dresden, Dresden; 3Helmholtz-Zentrum Dresden-Rossendorf, Institute of Radiooncology, Dresden; 4German Cancer Consortium (DKTK), D-01307 Dresden; 5German Cancer Research Center (DKFZ), D-69120 Heidelberg, Germany Received March 31, 2015; Accepted May 4, 2015 DOI: 10.3892/or.2015.4006 Abstract. Epidermal growth factor receptor (EGFR) signaling Introduction plays an important role in tumor cell resistance to therapy. In addition to ligand binding, mutual and cooperative interac- Epidermal growth factor receptor (EGFR) signaling is known tions of EGFR with integrin cell adhesion receptors critically to be deregulated in many human tumors (1,2). Causative are influence proper downstream signaling through a number of EGFR gene amplifications and mutations resulting in receptor bridging adapter proteins. In the present study, we analyzed overexpression and constitutively active EGFR tyrosine the role of two of these adapter proteins, called PINCH1 kinase activation. Due to its substantial role in -

Role of Phytochemicals in Colon Cancer Prevention: a Nutrigenomics Approach
Role of phytochemicals in colon cancer prevention: a nutrigenomics approach Marjan J van Erk Promotor: Prof. Dr. P.J. van Bladeren Hoogleraar in de Toxicokinetiek en Biotransformatie Wageningen Universiteit Co-promotoren: Dr. Ir. J.M.M.J.G. Aarts Universitair Docent, Sectie Toxicologie Wageningen Universiteit Dr. Ir. B. van Ommen Senior Research Fellow Nutritional Systems Biology TNO Voeding, Zeist Promotiecommissie: Prof. Dr. P. Dolara University of Florence, Italy Prof. Dr. J.A.M. Leunissen Wageningen Universiteit Prof. Dr. J.C. Mathers University of Newcastle, United Kingdom Prof. Dr. M. Müller Wageningen Universiteit Dit onderzoek is uitgevoerd binnen de onderzoekschool VLAG Role of phytochemicals in colon cancer prevention: a nutrigenomics approach Marjan Jolanda van Erk Proefschrift ter verkrijging van graad van doctor op gezag van de rector magnificus van Wageningen Universiteit, Prof.Dr.Ir. L. Speelman, in het openbaar te verdedigen op vrijdag 1 oktober 2004 des namiddags te vier uur in de Aula Title Role of phytochemicals in colon cancer prevention: a nutrigenomics approach Author Marjan Jolanda van Erk Thesis Wageningen University, Wageningen, the Netherlands (2004) with abstract, with references, with summary in Dutch ISBN 90-8504-085-X ABSTRACT Role of phytochemicals in colon cancer prevention: a nutrigenomics approach Specific food compounds, especially from fruits and vegetables, may protect against development of colon cancer. In this thesis effects and mechanisms of various phytochemicals in relation to colon cancer prevention were studied through application of large-scale gene expression profiling. Expression measurement of thousands of genes can yield a more complete and in-depth insight into the mode of action of the compounds. -

UNIVERSITY of CALIFORNIA RIVERSIDE Investigations Into The
UNIVERSITY OF CALIFORNIA RIVERSIDE Investigations into the Role of TAF1-mediated Phosphorylation in Gene Regulation A Dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Cell, Molecular and Developmental Biology by Brian James Gadd December 2012 Dissertation Committee: Dr. Xuan Liu, Chairperson Dr. Frank Sauer Dr. Frances M. Sladek Copyright by Brian James Gadd 2012 The Dissertation of Brian James Gadd is approved Committee Chairperson University of California, Riverside Acknowledgments I am thankful to Dr. Liu for her patience and support over the last eight years. I am deeply indebted to my committee members, Dr. Frank Sauer and Dr. Frances Sladek for the insightful comments on my research and this dissertation. Thanks goes out to CMDB, especially Dr. Bachant, Dr. Springer and Kathy Redd for their support. Thanks to all the members of the Liu lab both past and present. A very special thanks to the members of the Sauer lab, including Silvia, Stephane, David, Matt, Stephen, Ninuo, Toby, Josh, Alice, Alex and Flora. You have made all the years here fly by and made them so enjoyable. From the Sladek lab I want to thank Eugene, John, Linh and Karthi. Special thanks go out to all the friends I’ve made over the years here. Chris, Amber, Stephane and David, thank you so much for feeding me, encouraging me and keeping me sane. Thanks to the brothers for all your encouragement and prayers. To any I haven’t mentioned by name, I promise I haven’t forgotten all you’ve done for me during my graduate years. -

The Role of Cdep in the Embryonic Morphogenesis of Drosophila Melanogaster
The Role of Cdep in the Embryonic Morphogenesis of Drosophila melanogaster Dissertation zur Erlangung des akademischen Grades Doctor rerum naturalium (Dr. rer.nat.) vorgelegt der Fakultät Mathematik und Naturwissenschaften der Technischen Universität Dresden von Anne Morbach Geboren am 05. August 1984 in Nordhausen, Deutschland Eingereicht am 30. Oktober 2015 Die Dissertation wurde in der Zeit von April 2012 bis Oktober 2015 im Max-Planck-Institut für molekulare Zellbiologie und Genetik angefertigt Gutachter: Prof. Dr. Elisabeth Knust Prof. Dr. Christian Dahmann Now, here, you see, it takes all the running you can do, to keep in the same place. Lewis Carroll, Through the Looking-Glass I Contents List of Figures V List of Tables VII Abstract IX List of Abbreviations XI 1 Introduction 1 1.1 Epithelial cell polarity . 1 1.1.1 Cellularization and formation of the primary epithelium . 1 1.1.1.1 Establishment of epithelial polarity and adhesion . 2 1.1.2 The epithelial polarity network . 3 1.1.3 Cell-cell adhesion . 5 1.1.3.1 Adherens junctions . 5 1.1.3.2 Septate junctions . 6 1.2 Epithelial movements in Drosophila embryonic morphogenesis . 6 1.2.1 Epithelial tube formation during Drosophila embryogenesis . 7 1.2.2 Coordinated migration of epithelial sheets during Dros. embryogenesis 7 1.2.2.1 FERM domain proteins in epithelial migration . 9 1.2.2.2 Cdep . 10 1.3 Mutagenesis with the CRISPR/Cas9 system . 12 2 Aim of My PhD Thesis Work 15 3 Preliminary Work 17 4 Results 19 4.1 A screen for novel regulators in Drosophila embryonic morphogenesis . -

Anti-NCK2 Antibody (ARG59810)
Product datasheet [email protected] ARG59810 Package: 100 μl anti-NCK2 antibody Store at: -20°C Summary Product Description Rabbit Polyclonal antibody recognizes NCK2 Tested Reactivity Hu, Ms, Rat Tested Application IHC-P, WB Host Rabbit Clonality Polyclonal Isotype IgG Target Name NCK2 Antigen Species Human Immunogen Recombinant fusion protein corresponding to aa. 1-380 of Human NCK2 (NP_003572.2). Conjugation Un-conjugated Alternate Names Nck-2; NCKbeta; NCK adaptor protein 2; SH2/SH3 adaptor protein NCK-beta; GRB4; Growth factor receptor-bound protein 4; Cytoplasmic protein NCK2 Application Instructions Application table Application Dilution IHC-P 1:50 - 1:200 WB 1:500 - 1:2000 Application Note * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. Positive Control Rat brain, Mouse lung and 293T Calculated Mw 43 kDa Observed Size 43 kDa Properties Form Liquid Purification Affinity purified. Buffer PBS (pH 7.3), 0.02% Sodium azide and 50% Glycerol. Preservative 0.02% Sodium azide Stabilizer 50% Glycerol Storage instruction For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C. Storage in frost free freezers is not recommended. Avoid repeated freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use. www.arigobio.com 1/2 Note For laboratory research only, not for drug, diagnostic or other use. Bioinformation Gene Symbol NCK2 Gene Full Name NCK adaptor protein 2 Background This gene encodes a member of the NCK family of adaptor proteins. -

By Submitted in Partial Satisfaction of the Requirements for Degree of in In
BCL6 maintains thermogenic capacity of brown adipose tissue during dormancy by Vassily Kutyavin DISSERTATION Submitted in partial satisfaction of the requirements for degree of DOCTOR OF PHILOSOPHY in Biomedical Sciences in the GRADUATE DIVISION of the UNIVERSITY OF CALIFORNIA, SAN FRANCISCO Approved: ______________________________________________________________________________Eric Verdin Chair ______________________________________________________________________________Ajay Chawla ______________________________________________________________________________Ethan Weiss ______________________________________________________________________________ ______________________________________________________________________________ Committee Members Copyright 2019 by Vassily Kutyavin ii Dedicated to everyone who has supported me during my scientific education iii Acknowledgements I'm very grateful to my thesis adviser, Ajay Chawla, for his mentorship and support during my dissertation work over the past five years. Throughout my time in his lab, I was always able to rely on his guidance, and his enthusiasm for science was a great source of motivation. Even when he was traveling, he could easily be reached for advice by phone or e- mail. I am particularly grateful for his help with writing the manuscript, which was probably the most challenging aspect of graduate school for me. I am also very grateful to him for helping me find a postdoctoral fellowship position. Ajay's inquisitive and fearless approach to science have been a great inspiration to me. In contrast to the majority of scientists who focus narrowly on a specific topic, Ajay pursued fundamental questions across a broad range of topics and was able to make tremendous contributions. My experience in his lab instilled in me a deep appreciation for thinking about the entire organism from an evolutionary perspective and focusing on the key questions that escape the attention of the larger scientific community. As I move forward in my scientific career, there is no doubt that I will rely on him as a role model. -

Modular and Distinct Plexin-A4/FARP2/Rac1 Signaling Controls Dendrite Morphogenesis
The Journal of Neuroscience, July 8, 2020 • 40(28):5413–5430 • 5413 Development/Plasticity/Repair Modular and Distinct Plexin-A4/FARP2/Rac1 Signaling Controls Dendrite Morphogenesis Victor Danelon,1* Ron Goldner,2* Edward Martinez,1 Irena Gokhman,2 Kimberly Wang,1 Avraham Yaron,2 and Tracy S. Tran1 1Department of Biological Sciences, Rutgers University, Newark, New Jersey 07102, and 2Department of Biomolecular Sciences, Weizmann Institute of Science, Rehovot, 76100, Israel Diverse neuronal populations with distinct cellular morphologies coordinate the complex function of the nervous system. Establishment of distinct neuronal morphologies critically depends on signaling pathways that control axonal and dendritic devel- opment. The Sema3A-Nrp1/PlxnA4 signaling pathway promotes cortical neuron basal dendrite arborization but also repels axons. However, the downstream signaling components underlying these disparate functions of Sema3A signaling are unclear. Using the KRK-AAA novel PlxnA4 knock-in male and female mice, generated by CRISPR/cas9, we show here that the KRK motif in the PlxnA4 cytoplasmic domain is required for Sema3A-mediated cortical neuron dendritic elaboration but is dispensable for inhibitory axon guidance. The RhoGEF FARP2, which binds to the KRK motif, shows identical functional specificity as the KRK motif in the PlxnA4 receptor. We find that Sema3A activates the small GTPase Rac1, and that Rac1 activity is required for dendrite elaboration but not axon growth cone collapse. This work identifies a novel Sema3A-Nrp1/PlxnA4/FARP2/Rac1 signaling pathway that specifi- cally controls dendritic morphogenesis but is dispensable for repulsive guidance events. Overall, our results demonstrate that the divergent signaling output from multifunctional receptor complexes critically depends on distinct signaling motifs, highlighting the modular nature of guidance cue receptors and its potential to regulate diverse cellular responses. -

Stroke Genetics and Genomics
Faculdade de Medicina da Universidade de Lisboa Unidade Neurológica de Investigação Clínica PhD Thesis Stroke Genetics and Genomics Tiago Krug Coelho Host Institution: Instituto Gulbenkian de Ciência Supervisor at Instituto Gulbenkian de Ciência: Doctor Sofia Oliveira Supervisor at Faculdade de Medicina da Universidade de Lisboa: Professor José Ferro PhD in Biomedical Sciences Specialization in Neurosciences 2010 Stroke Genetics and Genomics A ciência tem, de facto, um único objectivo: a verdade. Não esgota perfeitamente a sua tarefa se não descobre a causa do todo. Chiara Lubich i Stroke Genetics and Genomics ii Stroke Genetics and Genomics A impressão desta dissertação foi aprovada pela Comissão Coordenadora do Conselho Científico da Faculdade de Medicina de Lisboa em reunião de 28 de Setembro de 2010. iii Stroke Genetics and Genomics iv Stroke Genetics and Genomics As opiniões expressas são da exclusiva responsabilidade do seu autor. v Stroke Genetics and Genomics vi Stroke Genetics and Genomics Abstract ABSTRACT This project presents a comprehensive approach to the identification of new genes that influence the risk for developing stroke. Stroke is the leading cause of death in Portugal and the third leading cause of death in the developed world. It is even more disabling than lethal, and the persistent neurological impairment and physical disability caused by stroke have a very high socioeconomic cost. Moreover, the number of affected individuals is expected to increase with the current aging of the population. Stroke is a “brain attack” cutting off vital blood and oxygen to the brain cells and it is a complex disease resulting from environmental and genetic factors. -

Mitochondrial Genetics
Mitochondrial genetics Patrick Francis Chinnery and Gavin Hudson* Institute of Genetic Medicine, International Centre for Life, Newcastle University, Central Parkway, Newcastle upon Tyne NE1 3BZ, UK Introduction: In the last 10 years the field of mitochondrial genetics has widened, shifting the focus from rare sporadic, metabolic disease to the effects of mitochondrial DNA (mtDNA) variation in a growing spectrum of human disease. The aim of this review is to guide the reader through some key concepts regarding mitochondria before introducing both classic and emerging mitochondrial disorders. Sources of data: In this article, a review of the current mitochondrial genetics literature was conducted using PubMed (http://www.ncbi.nlm.nih.gov/pubmed/). In addition, this review makes use of a growing number of publically available databases including MITOMAP, a human mitochondrial genome database (www.mitomap.org), the Human DNA polymerase Gamma Mutation Database (http://tools.niehs.nih.gov/polg/) and PhyloTree.org (www.phylotree.org), a repository of global mtDNA variation. Areas of agreement: The disruption in cellular energy, resulting from defects in mtDNA or defects in the nuclear-encoded genes responsible for mitochondrial maintenance, manifests in a growing number of human diseases. Areas of controversy: The exact mechanisms which govern the inheritance of mtDNA are hotly debated. Growing points: Although still in the early stages, the development of in vitro genetic manipulation could see an end to the inheritance of the most severe mtDNA disease. Keywords: mitochondria/genetics/mitochondrial DNA/mitochondrial disease/ mtDNA Accepted: April 16, 2013 Mitochondria *Correspondence address. The mitochondrion is a highly specialized organelle, present in almost all Institute of Genetic Medicine, International eukaryotic cells and principally charged with the production of cellular Centre for Life, Newcastle energy through oxidative phosphorylation (OXPHOS).