Visualization Methods for Molecular Studies on the Web Platform
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Web-Based 3D Molecular Structure Editor and Visualizer Platform
Mohebifar and Sajadi J Cheminform (2015) 7:56 DOI 10.1186/s13321-015-0101-7 SOFTWARE Open Access Chemozart: a web‑based 3D molecular structure editor and visualizer platform Mohamad Mohebifar* and Fatemehsadat Sajadi Abstract Background: Chemozart is a 3D Molecule editor and visualizer built on top of native web components. It offers an easy to access service, user-friendly graphical interface and modular design. It is a client centric web application which communicates with the server via a representational state transfer style web service. Both client-side and server-side application are written in JavaScript. A combination of JavaScript and HTML is used to draw three-dimen- sional structures of molecules. Results: With the help of WebGL, three-dimensional visualization tool is provided. Using CSS3 and HTML5, a user- friendly interface is composed. More than 30 packages are used to compose this application which adds enough flex- ibility to it to be extended. Molecule structures can be drawn on all types of platforms and is compatible with mobile devices. No installation is required in order to use this application and it can be accessed through the internet. This application can be extended on both server-side and client-side by implementing modules in JavaScript. Molecular compounds are drawn on the HTML5 Canvas element using WebGL context. Conclusions: Chemozart is a chemical platform which is powerful, flexible, and easy to access. It provides an online web-based tool used for chemical visualization along with result oriented optimization for cloud based API (applica- tion programming interface). JavaScript libraries which allow creation of web pages containing interactive three- dimensional molecular structures has also been made available. -

Chemdoodle Web Components: HTML5 Toolkit for Chemical Graphics, Interfaces, and Informatics Melanie C Burger1,2*
Burger. J Cheminform (2015) 7:35 DOI 10.1186/s13321-015-0085-3 REVIEW Open Access ChemDoodle Web Components: HTML5 toolkit for chemical graphics, interfaces, and informatics Melanie C Burger1,2* Abstract ChemDoodle Web Components (abbreviated CWC, iChemLabs, LLC) is a light-weight (~340 KB) JavaScript/HTML5 toolkit for chemical graphics, structure editing, interfaces, and informatics based on the proprietary ChemDoodle desktop software. The library uses <canvas> and WebGL technologies and other HTML5 features to provide solutions for creating chemistry-related applications for the web on desktop and mobile platforms. CWC can serve a broad range of scientific disciplines including crystallography, materials science, organic and inorganic chemistry, biochem- istry and chemical biology. CWC is freely available for in-house use and is open source (GPL v3) for all other uses. Keywords: ChemDoodle Web Components, Chemical graphics, Animations, Cheminformatics, HTML5, Canvas, JavaScript, WebGL, Structure editor, Structure query Introduction Mobile browsers did support HTML5, which opened How we communicate chemical information is increas- the door to web applications built with only HTML, ingly technology driven. Learning management systems, CSS and JavaScript (JS), such as the ChemDoodle Web virtual classrooms and MOOCs are a few examples where Components. chemistry educators need forward compatible tools for digital natives. Companies that implement emerg- Review ing web technologies can find efficiencies and benefit The ChemDoodle Web Components technology stack from competitive advantages. The first chemical graph- and features ics toolkit for the web, MDL Chime, was introduced in The ChemDoodle Web Components library, released in 1996 [1]. Based on the molecular visualization program 2009, is the first chemistry toolkit for structure viewing RasMol, Chime was developed as a plugin for Netscape and editing that is originally built using only web stand- and later for Internet Explorer and Firefox. -

Water and Salt at the Lipid-Solvent Interface
University of South Florida Scholar Commons Graduate Theses and Dissertations Graduate School April 2019 Water and Salt at the Lipid-Solvent Interface James M. Kruczek University of South Florida, [email protected] Follow this and additional works at: https://scholarcommons.usf.edu/etd Part of the Physics Commons Scholar Commons Citation Kruczek, James M., "Water and Salt at the Lipid-Solvent Interface" (2019). Graduate Theses and Dissertations. https://scholarcommons.usf.edu/etd/8380 This Dissertation is brought to you for free and open access by the Graduate School at Scholar Commons. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Scholar Commons. For more information, please contact [email protected]. Water and Salt at the Lipid-Solvent Interface by James M. Kruczek A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Applied Physics Department of Physics College of Arts and Sciences University of South Florida Major Professor: Sagar A. Pandit, Ph.D. Ullah, Ghanim, Ph.D. Robert S. Hoy, Ph.D. Jianjun Pan, Ph.D. Yicheng Tu, Ph.D. Date of Approval: March 26, 2019 Keywords: Lipid Bilayer, Ionic Solvents, Ether Lipids, Molecular Simulations Copyright ⃝c 2018, James M. Kruczek Dedication To my wife Nicole, without whom none of this would be possible. To my father Michael, who labored for his family till his last days. To my family, for all of their support. Acknowledgments The work presented in this document would not be possible without the assistance of many academic professionals. In particular, I would like to acknowledge my major advisor Dr. -

Tracking of Bacterial Metabolism with Azidated Precursors and Click- Chemistry”
Tracking of bacterial metabolism with azidated precursors and click-chemistry Dissertation zur Erlangung des Doktorgrades der Naturwissenschaften vorgelegt beim Fachbereich für Biowissenschaften (15) der Johann Wolfang Goethe Universität in Frankfurt am Main von Alexander J. Pérez aus Nürnberg Frankfurt am Main 2015 Dekanin: Prof. Dr. Meike Piepenbring Gutachter: Prof. Dr. Helge B. Bode Zweitgutachter: Prof. Dr. Joachim W. Engels Datum der Disputation: 2 Danksagung Ich danke meinen Eltern für die stete und vielseitige Unterstützung, deren Umfang ich sehr zu schätzen weiß. Herrn Professor Dr. Helge B. Bode gilt mein besonderer Dank für die Übernahme als Doktorand und für die Gelegenheit meinen Horizont in diesen mich stets faszinierenden Themenbereich in dieser Tiefe erweitern zu lassen. Seine persönliche und fachliche Unterstützung bei der Projektwahl und der entsprechenden Umsetzung ist in dieser Form eine Seltenheit und ich bin mir dieser Tatsache voll bewusst. Gerade die zusätzlich erworbenen Kenntnisse im Bereich der Biologie, sowie der Wert interdisziplinärer Zusammenarbeit ist mir durch zahlreiche freundliche und wertvolle Mitglieder der Arbeitsgruppe bewusst geworden und viele zündenden Ideen wären ohne sie womöglich nie aufgekommen. Einen besonderen Dank möchte ich in diesem Kontext Wolfram Lorenzen und Sebastian Fuchs, die gerade in der Anfangszeit eine große Hilfe waren, ausdrücken. Dies gilt ebenso für die „N100-Crew“ und sämtliche Freunde, die in dieser Zeit zu mir standen und diesen Lebensabschnitt unvergesslich gemacht haben. -

Titelei 1..30
Johann Gasteiger, Thomas Engel (Eds.) Chemoinformatics Chemoinformatics: A Textbook. Edited by Johann Gasteiger and Thomas Engel Copyright 2003 Wiley-VCH Verlag GmbH & Co. KGaA. ISBN: 3-527-30681-1 Related Titles from WILEY-VCH Hans-Dieter HoÈltje, Wolfgang Sippl, Didier Rognan, Gerd Folkers Molecular Modeling 2003 ISBN 3-527-30589-0 Helmut GuÈnzler, Hans-Ulrich Gremlich IR Spectroscopy An Introduction 2002 ISBN 3-527-28896-1 Jure Zupan, Johann Gasteiger Neural Networks in Chemistry and Drug Design An Introduction 1999 ISBN 3-527-29778-2 (HC) ISBN 3-527-29779-0 (SC) Siegmar Braun, Hans-Otto Kalinowski, Stefan Berger 150 and More Basic NMR Experiments A Practical Course 1998 ISBN 3-527-29512-7 Johann Gasteiger, Thomas Engel (Eds.) Chemoinformatics A Textbook Editors: This book was carefully produced. Never- theless, editors, authors and publisher do Prof. Dr. Johann Gasteiger not warrant the information contained Computer-Chemie-Centrum and Institute therein to be free of errors. Readers are ofOrganic Chemistry advised to keep in mind that statements, University ofErlangen-NuÈrnberg data, illustrations, procedural details or NaÈgelsbachstraûe 25 other items may inadvertently be inaccurate. 91052 Erlangen Germany Library of Congress Card No.: applied for A catalogue record for this book is available from the British Library. Dr. Thomas Engel Computer-Chemie-Centrum and Institute Bibliographic information published by ofOrganic Chemistry Die Deutsche Bibliothek University ofErlangen-NuÈrnberg Die Deutsche Bibliothek lists this publica- NaÈgelsbachstraûe 25 tion in the Deutsche Nationalbibliografie; 91052 Erlangen detailed bibliographic data is available in the Germany Internet at http://dnb.ddb.de ISBN 3-527-30681-1 c 2003 WILEY-VCH Verlag GmbH & Co. -
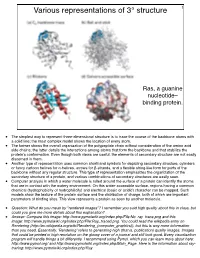
Various Representations of 3° Structure 1
Various representations of 3° structure 1 Ras, a guanine nucleotide– binding protein. • The simplest way to represent three-dimensional structure is to trace the course of the backbone atoms with a solid line; the most complex model shows the location of every atom. • The former shows the overall organization of the polypeptide chain without consideration of the amino acid side chains; the latter details the interactions among atoms that form the backbone and that stabilize the protein’s conformation. Even though both views are useful, the elements of secondary structure are not easily discerned in them. • Another type of representation uses common shorthand symbols for depicting secondary structure, cylinders or fancy cartoon helices for α-helices, arrows for β-strands, and a flexible string-like form for parts of the backbone without any regular structure. This type of representation emphasizes the organization of the secondary structure of a protein, and various combinations of secondary structures are easily seen. • Computer analysis in which a water molecule is rolled around the surface of a protein can identify the atoms that are in contact with the watery environment. On this water-accessible surface, regions having a common chemical (hydrophobicity or hydrophilicity) and electrical (basic or acidic) character can be mapped. Such models show the texture of the protein surface and the distribution of charge, both of which are important parameters of binding sites. This view represents a protein as seen by another molecule. • Question: What do you mean by "rendered images"? I remember you said high quality about this in class, but could you give me more details about this explanation? • Answer: Compare this image: http://www.pymolwiki.org/index.php/File:No_ray_trace.png and this image: http://www.pymolwiki.org/index.php/File:Ray_traced.png. -

Drug Discovery Informatics Registration Systems and Underlying Toolkits (Appendices 1 and 2) 271 Subject Appendices
SUBJECT APPENDICES DRUG DISCOVERY INFORMATICS REGISTRATION SYSTEMS AND UNDERLYING TOOLKITS (APPENDICES 1 AND 2) Appendix 1: DRUG, MOLECULAR REGISTRATION SYSTEMS, AND CHEMISTRY DATA CARTRIDGES Product Company Page Website Isentris, MDL- MDL 171 http://www.mdli.com Base, ISIS- Base ChemOffice Cambridgesoft 73 http://www.cambridgesoft.com Cambridgesoft CambridgeSoft 73 http://www.cambridgesoft.com/solutions/ Enterprise Solutions ActivityBase IDBS 152 http://www.idbs.com/products/abase/ Accord Accelrys 51 http://www.accelrys.com AUSPYX Tripos 261 http://www.tripos.com CBIS ChemInnovation 103 http://www.cheminnovation.com/ (Chemical and Software Biological Informatics Systems) ACD/SpecDB ACD Labs 59 http://www.acdlabs.com (Spectral Data Management System) - Analytical data capture tools Super Package BioByte 71 http://www.biobyte.com/index.html /THOR (Daylight) JChem ChemAxon 89 http://www.chemaxon.com Cartridge Software for Infochem 156 http://www.infochem.de/en/company/ Chemical index.shtml databases and chemical data processes 271 272 CHEMOINFORMATICS The InfoChem Infochem 156 http://www.infochem.de/eng/index.htm Chemistry Cartridge for Oracle Molecular Collaborative 115 http://www.collaborativedrug.com/ Databank Drug Discovery DayCart Daylight 123 http://www.daylight.com/ Chemical Information Systems, Inc. CHED, TimTec 254 http://software.timtec.net/ Chemdbsoft MDL Isentris MDL 171 http://www.mdli.com platform (ISIS Base), ChemBio AE, AssayExplorer Appendix 2: CHEMOINFORMATICS TOOLKITS TO DEVELOP APPLICATIONS Product Company Page Website -

Visualizing Protein Structures-Tools and Trends
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 12 January 2020 Visualizing protein structures - tools and trends 1,2 3 1,2 X. Martinez , M. Chavent , M. Baaden 1) CNRS, Université de Paris, UPR 9080, Laboratoire de Biochimie Théorique, 13 rue Pierre et Marie Curie, F-75005, Paris, France 2) Institut de Biologie Physico-Chimique-Fondation Edmond de Rotschild, PSL Research University, Paris, France 3) Institut de Pharmacologie et de Biologie Structurale IPBS, Université de Toulouse, CNRS, UPS, Toulouse, France Abstract Molecular visualisation is fundamental in the current scientific literature, textbooks and dissemination materials, forming an essential support for presenting results, reasoning on and formulating hypotheses related to molecular structure. Visual exploration has become easily accessible on a broad variety of platforms thanks to advanced software tools that render a great service to the scientific community. These tools are often developed across disciplines bridging computer science, biology and chemistry. Here we first describe a few Swiss Army knives geared towards protein visualisation for everyday use with an existing large user base, then focus on more specialised tools for peculiar needs that are not yet as broadly known. Our selection is by no means exhaustive, but reflects a diverse snapshot of scenarios that we consider informative for the reader. We end with an account of future trends and perspectives. Keywords Molecular Graphics, Protein visualization, Software tools, Virtual reality Introduction Many parts of science rely on a visualization-driven cycle of experimentation, reasoning, conjecture and validation, even more so in relation with structural biology and biophysics. Molecular visualization (1) in particular is now broadly used in many contexts, with the purpose of illustration in the scientific literature or the aim to gain insight about primary research data. -

Engine for Molecule Visualization in a Web Browser
MASARYKOVA UNIVERZITA FAKULTA}w¡¢£¤¥¦§¨ INFORMATIKY !"#$%&'()+,-./012345<yA| Engine for Molecule Visualization in a Web Browser MASTER’S THESIS Jaromír Svoboda Brno, spring 2014 Declaration Hereby I declare, that this paper is my original authorial work, which I have worked out by my own. All sources, references and literature used or excerpted during elaboration of this work are properly cited and listed in complete reference to the due source. Jaromír Svoboda Advisor: RNDr. David Sehnal ii Acknowledgement I would like to thank my supervisor RNDr. David Sehnal for patient guidance and informed advice throughout writing this thesis. iii Abstract The main focus of this master’s thesis is the design and implementa- tion of lightweight molecular visualization engine (called LiveMol) in form of a JavaScript library. The engine utilizes widely adopted WebGL API to display GPU-accelerated graphics in web browsers. Due to the size of complex protein molecules, the primary goal is high performance. Furthermore, the design of LiveMol enables users to extend the core functionality by simply defining custom coloring schemes of molecule models or implement completely new visual- ization modes. iv Keywords molecular visualization, protein, secondary structure, WebGL, LiveMol v Contents I INTRODUCTION 1 1 Introduction ............................2 II CURRENT DEVELOPMENTS 3 2 Current Molecular Visualization Software ..........4 2.1 Desktop Applications ....................4 2.1.1 Visual Molecular Dynamic . .4 2.1.2 PyMOL . .4 2.1.3 RasMol/OpenRasMol . .5 2.1.4 BALL/BALLView . .5 2.1.5 Gabedit . .5 2.1.6 QuteMol . .5 2.1.7 Avogadro . .6 2.2 JavaScript-based Web Applications ............6 2.2.1 Jmol/JSmol . -

Illustrative Molecular Visualization with Continuous Abstraction Matthew Van Der Zwan, Wouter Lueks, Henk Bekker, Tobias Isenberg
Illustrative Molecular Visualization with Continuous Abstraction Matthew van der Zwan, Wouter Lueks, Henk Bekker, Tobias Isenberg To cite this version: Matthew van der Zwan, Wouter Lueks, Henk Bekker, Tobias Isenberg. Illustrative Molecular Visu- alization with Continuous Abstraction. Computer Graphics Forum, Wiley, 2011, 30 (3), pp.683-690. 10.1111/j.1467-8659.2011.01917.x. hal-00781508 HAL Id: hal-00781508 https://hal.inria.fr/hal-00781508 Submitted on 27 Jan 2013 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Public Domain Eurographics / IEEE Symposium on Visualization 2011 (EuroVis 2011) Volume 30 (2011), Number 3 H. Hauser, H. Pfister, and J. J. van Wijk (Guest Editors) Illustrative Molecular Visualization with Continuous Abstraction Matthew van der Zwan,1 Wouter Lueks,1 Henk Bekker,1 and Tobias Isenberg1,2 1Johann Bernoulli Institute of Mathematics and Computer Science, University of Groningen, The Netherlands 2DIGITEO in collaboration with VENISE–LIMSI–CNRS and AVIZ–INRIA, Saclay, France Abstract Molecular systems may be visualized with various degrees of structural abstraction, support of spatial perception, and ‘illustrativeness.’In this work we propose and realize methods to create seamless transformations that allow us to affect and change each of these three parameters individually. -

Free Download Plugin for Firefox Internet Explorer
Free download plugin for firefox internet explorer click here to download of add-ons available here, download Mozilla Firefox, a fast, free way to surf the Web! Not only an enhanced IE Tab, but also an enhanced Internet Explorer with all plugins except Adobe Flash in the official x64 version, which breaks Fire IE. /RemoveFx41x64PluginRestrict/patch4fx45xz/download. Open in IE™ addon gives you the ability to open any link in an Internet Explorer Follow the instruction in the above GitHub page to download and install the. You need to download Firefox to install this add-on. About this extension. Just like the classic IE Tab, this updated version of IE Tab allows you to use IE to. The Office Web Apps Browser Plugin installs an add-on that enables Office documents to be opened directly from Firefox into the appropriate. If you find yourself doing the same thing even though Firefox is your browser of choice, try IE Tab for Firefox. This clever extension lets you open IE-only Web. The Google Earth plug-in for Firefox and Internet Explorer allows users to view The Download Now link will prompt a local download of the Firefox extension. IE View - Open pages in Internet Explorer from Firefox. was v, available from the IE View Versions page at www.doorway.ru It is MPL-licensed, forkable, and free to modify and redistribute. IE. You'll find a great range of free browser plugins and free browser add ons at File Hippo. We are a trusted and official source of free programs to download. -

Bringing Molecular Dynamics Simulation Data Into View
Opinion Bringing Molecular Dynamics Simulation Data into View Peter W. Hildebrand,1,2,3,* Alexander S. Rose,4,@ and Johanna K.S. Tiemann1,@ Molecular dynamics (MD) simulations monitor time-resolved motions of macromolecules. While Highlights visualization of MD trajectories allows an instant and intuitive understanding of dynamics and MD simulations have become function, so far mainly static representations are provided in the published literature. Recent ad- routinely used over the past years vances in browser technology may allow for the sharing of trajectories through interactive visu- to investigate dynamic motions of alization on the web. We believe that providing intuitive and interactive visualization, along with macromolecules at the atomic related protocols and analysis data, promotes understanding, reliability, and reusability of MD level. simulations. Existing barriers for sharing MD simulations are discussed and emerging solutions Interactive visualization of MD tra- are highlighted. We predict that interactive visualization of MD trajectories will quickly be adop- jectories may provide an instant, ted by researchers, research consortiums, journals, and funding agencies to gather and transparent, and intuitive under- distribute results from MD simulations via the web. standing of complex dynamics. Dynamics Leads to Understanding Macromolecule Function Sharing of MD trajectories may generate transparency and trust, Molecular dynamics (MD,seeGlossary) simulations are a well-established technique to investigate allowing collaboration, knowledge time-resolved motions of biological macromolecules at atomic resolution [1,2]. Traditionally, macro- exchange, and data reuse. molecules such as enzymes, channels, transporters, or receptors have been perceived as being rigid entities mainly because structures obtained by X-ray crystallography are fixed in crystal lattices and Recent technological de- are therefore resolved as single, static snapshots.