Genetic Diversity, Phylogenetic Relationships and Conservation of Edwards's Pheasant Lophura Edwardsi
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Proposal for Amendment of Appendix I Or II for CITES Cop16
Original language: French CoP16 Prop. 17 CONVENTION ON INTERNATIONAL TRADE IN ENDANGERED SPECIES OF WILD FAUNA AND FLORA ____________________ Sixteenth meeting of the Conference of the Parties Bangkok (Thailand), 3-14 March 2013 CONSIDERATION OF PROPOSALS FOR AMENDMENT OF APPENDICES I AND II A. Proposal Delete Lophura imperialis from Appendix I and Amend the standard reference for birds adopted by the Conference of the Parties in the Annex to Resolution Conf. 12.11 (Rev. CoP15): "Dickinson, E. C. (ed.)(2003): The Howard and Moore Complete Checklist of the Birds of the World. Revised and enlarged 3rd Edition. 1039 pp. London (Christopher Helm)", inserting the following text in square brackets: [for all bird species – except for Lophura imperialis and the taxa mentioned below] B. Proponent Switzerland, as Depositary Government, at the request of the Animals Committee (proposal prepared by France)1. C. Supporting statement 1. Taxonomy 1.1 Class: Aves 1.2 Order: Galliformes 1.3 Family: Phasianidae 1.4 Genus, species, including author and year: Lophura imperialis, Delacour et Jabouille, 1924 Birdlife International no longer recognizes the imperial pheasant (Lophura imperialis) as a species (2011). According to the IUCN Species Survival Commission / World Pheasant Association Galliformes Specialist Group, Lophura imperialis is no longer a valid name and the taxon should be considered a natural hybrid between the species Lophura edwardsi (Oustalet, 1896) and Lophura nycthemera (Linné, 1758). As a result, Lophura imperialis has been removed from the IUCN Red List. 1 The geographical designations employed in this document do not imply the expression of any opinion whatsoever on the part of the CITES Secretariat or the United Nations Environment Programme concerning the legal status of any country, territory, or area, or concerning the delimitation of its frontiers or boundaries. -
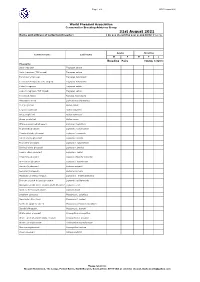
31St August 2021 Name and Address of Collection/Breeder: Do You Closed Ring Your Young Birds? Yes / No
Page 1 of 3 WPA Census 2021 World Pheasant Association Conservation Breeding Advisory Group 31st August 2021 Name and address of collection/breeder: Do you closed ring your young birds? Yes / No Adults Juveniles Common name Latin name M F M F ? Breeding Pairs YOUNG 12 MTH+ Pheasants Satyr tragopan Tragopan satyra Satyr tragopan (TRS ringed) Tragopan satyra Temminck's tragopan Tragopan temminckii Temminck's tragopan (TRT ringed) Tragopan temminckii Cabot's tragopan Tragopan caboti Cabot's tragopan (TRT ringed) Tragopan caboti Koklass pheasant Pucrasia macrolopha Himalayan monal Lophophorus impeyanus Red junglefowl Gallus gallus Ceylon junglefowl Gallus lafayettei Grey junglefowl Gallus sonneratii Green junglefowl Gallus varius White-crested kalij pheasant Lophura l. hamiltoni Nepal Kalij pheasant Lophura l. leucomelana Crawfurd's kalij pheasant Lophura l. crawfurdi Lineated kalij pheasant Lophura l. lineata True silver pheasant Lophura n. nycthemera Berlioz’s silver pheasant Lophura n. berliozi Lewis’s silver pheasant Lophura n. lewisi Edwards's pheasant Lophura edwardsi edwardsi Vietnamese pheasant Lophura e. hatinhensis Swinhoe's pheasant Lophura swinhoii Salvadori's pheasant Lophura inornata Malaysian crestless fireback Lophura e. erythrophthalma Bornean crested fireback pheasant Lophura i. ignita/nobilis Malaysian crestless fireback/Vieillot's Pheasant Lophura i. rufa Siamese fireback pheasant Lophura diardi Southern Cavcasus Phasianus C. colchicus Manchurian Ring Neck Phasianus C. pallasi Northern Japanese Green Phasianus versicolor -

Save Pdf (0.81
FFPS news The Oryx 100% Fund threat from the cage-bird trade in Trinidad. The second will be concerned with forest regenera- tion on the Tobago Main Ridge, which was Grants awarded devastated by Hurricane Flora in 1965. The team At its meeting on 7 June the FFPS Council will assess natural regeneration and compare it approved funding for the following projects. with growth in areas replanted with fast-growing exotic species to combat erosion. The results £800 for a project focusing on conservation of should be of wide interest in the Caribbean forest birds in Vietnam. The team consists of where forests are ever prone to hurricane damage i three people from the UK and two from Vietnam, (Project number 89/22/8). who will carry out surveys to discover good populations of Vietnamese pheasant Lophura £300 for the University of East Anglia Comoro hatinhensis and Edward's pheasant L.edwardsi Islands Expedition 1989. One British and three in areas suitable for protection as part of a five- Comorian people will reassess the status of year agreement with the Government of Viet- Livingstone's giant fruit bat Pteropus livingstonii nam. The team will also carry out preliminary following cyclone Calasanji in January 1989. surveys for orange-necked partridge Arboro- The team will also census the rare Grand Com- phila davidi, green peafowl Pavo muticus and oro scops owl Otus pauliani and the mongoose Germain's peacock-pheasant Polyplecton ger- lemur Lemur mongoz. Conservation recommen- maini. All these birds are considered to be dations and the possibility of a breeding pro- threatened (Project number 89/24/9). -

Vietnam Edwards's Pheasant Working Group Action Plan for The
Vietnam Edwards’s Pheasant Working Group Action Plan for the Conservation of the Edwards’s Pheasant Lophura edwardsi 2015 – 2020 Hanoi, March 2015 1 Table of content List of Acronyms .............................................................................................................................................. 3 Foreword ......................................................................................................................................................... 4 Executive Summary ......................................................................................................................................... 5 Introduction .................................................................................................................................................... 7 1. The Species.................................................................................................................................................. 9 1.1 Taxonomy and Ecology ......................................................................................................................... 9 1.2 Distribution: .......................................................................................................................................... 9 1.3 Habitat Requirements ......................................................................................................................... 11 1.4 Population Size and Trends ................................................................................................................ -

Colorado Birds the Colorado Field Ornithologists’ Quarterly
Vol. 45 No. 3 July 2011 Colorado Birds The Colorado Field Ornithologists’ Quarterly Remembering Abby Modesitt Chipping Sparrow Migration Sage-Grouse Spermatozoa Colorado Field Ornithologists PO Box 643, Boulder, Colorado 80306 www.cfo-link.org Colorado Birds (USPS 0446-190) (ISSN 1094-0030) is published quarterly by the Colo- rado Field Ornithologists, P.O. Box 643, Boulder, CO 80306. Subscriptions are obtained through annual membership dues. Nonprofit postage paid at Louisville, CO. POST- MASTER: Send address changes to Colorado Birds, P.O. Box 643, Boulder, CO 80306. Officers and Directors of Colorado Field Ornithologists: Dates indicate end of current term. An asterisk indicates eligibility for re-election. Terms expire 5/31. Officers: President: Jim Beatty, Durango, 2013; [email protected]; Vice President: Bill Kaempfer, Boulder, 2013*; [email protected]; Secretary: Larry Modesitt, Greenwood Village, 2013*; [email protected]; Treasurer: Maggie Boswell, Boulder, 2013; trea- [email protected] Directors: Lisa Edwards, Falcon, 2014*; Ted Floyd, Lafayette, 2014*; Brenda Linfield, Boulder, 2013*; Christian Nunes, Boulder, 2013*; Bob Righter, Denver, 2012*; Joe Roller, Denver, 2012* Colorado Bird Records Committee: Dates indicate end of current term. An asterisk indicates eligibility to serve another term. Terms expire 12/31. Chair: Larry Semo, Westminster; [email protected] Secretary: Doug Faulkner, Arvada (non-voting) Committee Members: John Drummond, Monument, 2013*; Peter Gent, Boulder, 2012; Bill Maynard, Colorado Springs, 2013; Bill Schmoker, Longmont, 2013*; David Silver- man, Rye, 2011*; Glenn Walbek, Castle Rock, 2012* Colorado Birds Quarterly: Editor: Nathan Pieplow, [email protected] Staff: Glenn Walbek (Photo Editor), [email protected]; Hugh Kingery (Field Notes Editor), [email protected]; Tony Leukering (In the Scope Editor), GreatGrayOwl@aol. -

Lophura Hatinhensis Is an Invalid Taxon
FORKTAIL 28 (2012): 129–135 Lophura hatinhensis is an invalid taxon ALAIN HENNACHE, SIMON P. MAHOOD, JONATHAN C. EAMES & ETTORE RANDI The Vietnamese Pheasant Lophura hatinhensis was described in 1975 from one male specimen which was superficially similar to Edwards’s Pheasant L. edwardsi but for four white (instead of dark metallic blue) tail feathers. Like L. edwardsi it is poorly known and highly threatened in the wild. Its status as a species has rarely been questioned despite its curious distribution and dubious morphological distinctiveness. To elucidate the taxonomic status of L. hatinhensis we examined the morphology of captive birds of both taxa and analysed mitochondrial DNA. These lines of evidence demonstrated that birds exhibiting the L. hatinhensis phenotype probably represent inbred L. edwardsi. Thus L. hatinhensis should be removed from the IUCN Red List and other checklists of valid extant bird species. Its apparent recent appearance alongside wild populations of L. edwardsi might be taken as evidence that wild populations of this species are also highly inbred and possibly close to extinction. INTRODUCTION each other’s closest relatives (Randi et al. 1997, Scott 1997, Hennache et al. 2003) and that they diverged within the last The Vietnamese Pheasant Lophura hatinhensis was described by 100,000 years (Scott 1997, Hennache et al. 1999). Although their Vo Quy (1975) in his book Chim Viet Nam (translation: ‘Birds phylogenetic relationships could not be accurately determined, Vietnam’) and has been widely recognised as a species ever since Scott (1997) proposed that they should be considered evolutionary (Sibley & Monroe 1990, Inskipp et al. 1996, BirdLife International significant units. -

Naamgeving Van Fazanten
Nomenclatuur Phasianini; wetenschappelijke, Nederlandse, Engelse, Duitse en Franse naamgeving Door: Han Assink I.s.m.: Peter Garson (chairman Pheasant Specialist Group), Alain Hennache (chairman EAZA - Pheasant TAG, chairman WPA-France), Heiner Jacken (bestuur WPA-Deutschland), Ludo Pinceel (docent biologie aan de Katholieke Hogeschool Kempen en lector aan de Katholieke Universiteit Leuven), Steven Vansteenkiste (curator vogels Zoo Antwerpen) en Ronald Wezeman (bestuur WPA-Benelux) World Pheasant Association – WPA-Benelux Nomenclatuur Phasianini (versie 1.0) Naamgeving van fazanten In het gebruik van fazantennamen komen veel verschillen voor. Deze brochure is bedoeld om mee te helpen om een standaard te maken voor naamgeving van fazanten. De bedoeling is om vertrekkende vanuit de wetenschappelijke namen het gebruik van namen in het Nederlands en in de andere talen Engels, Duits en Frans overzichtelijk te maken. Soms komt het voor dat namen overgenomen worden uit andere talen, maar dat de schrijfwijze enigszins afwijkt. Voorbeelden hiervan in het Nederlands / Vlaams zijn bijvoorbeeld: tragopan of tragopaan. In dit overzicht hebben wij gekozen voor tragopaan. Het oudere woord saterhoen wordt niet of nauwelijks nog gebruikt in de spreektaal van kwekers en wetenschappers. Bovendien zijn er Aziatische landen die er de voorkeur aan geven om de naamgeving van een fazant aan te passen aan deze tijd. Een goed voorbeeld hiervan is de bergpauwfazant, die voorheen en nu nog vaak als Rothschildpauwfazant werd aangeduid. Het document is slechts een -

Phasianinae Species Tree
Phasianinae Blood Pheasant,Ithaginis cruentus Ithaginini ?Western Tragopan, Tragopan melanocephalus Satyr Tragopan, Tragopan satyra Blyth’s Tragopan, Tragopan blythii Temminck’s Tragopan, Tragopan temminckii Cabot’s Tragopan, Tragopan caboti Lophophorini ?Snow Partridge, Lerwa lerwa Verreaux’s Monal-Partridge, Tetraophasis obscurus Szechenyi’s Monal-Partridge, Tetraophasis szechenyii Chinese Monal, Lophophorus lhuysii Himalayan Monal, Lophophorus impejanus Sclater’s Monal, Lophophorus sclateri Koklass Pheasant, Pucrasia macrolopha Wild Turkey, Meleagris gallopavo Ocellated Turkey, Meleagris ocellata Tetraonini Ruffed Grouse, Bonasa umbellus Hazel Grouse, Tetrastes bonasia Chinese Grouse, Tetrastes sewerzowi Greater Sage-Grouse / Sage Grouse, Centrocercus urophasianus Gunnison Sage-Grouse / Gunnison Grouse, Centrocercus minimus Dusky Grouse, Dendragapus obscurus Sooty Grouse, Dendragapus fuliginosus Sharp-tailed Grouse, Tympanuchus phasianellus Greater Prairie-Chicken, Tympanuchus cupido Lesser Prairie-Chicken, Tympanuchus pallidicinctus White-tailed Ptarmigan, Lagopus leucura Rock Ptarmigan, Lagopus muta Willow Ptarmigan / Red Grouse, Lagopus lagopus Siberian Grouse, Falcipennis falcipennis Spruce Grouse, Canachites canadensis Western Capercaillie, Tetrao urogallus Black-billed Capercaillie, Tetrao parvirostris Black Grouse, Lyrurus tetrix Caucasian Grouse, Lyrurus mlokosiewiczi Tibetan Partridge, Perdix hodgsoniae Gray Partridge, Perdix perdix Daurian Partridge, Perdix dauurica Reeves’s Pheasant, Syrmaticus reevesii Copper Pheasant, Syrmaticus -

AC26 Doc. 13.2.1, Annex – 1 Species: Argusianus Argus
AC26 Doc. 13.2.1 Annex English only / únicamente en inglés / seulement en anglais WORLD PHEASANT ASSOCIATION - CITES GALLIFORMES PERIODIC REVIEW FINAL REPORT The approach used by the World Pheasant Association in the preparation of this report was to: Outline information gathered about the species considered for evaluation in this review was first drawn from consolidated sources, with the intent of providing a baseline summary. Consequently, only a small number of base references (containing the most current information) were referred to. The term ‘Information is not readily available’ used in sections throughout the document, indicates that the sources consulted did not contain such information and, therefore, additional information may be available either in the literature or from informed sources., such as range states Draft accounts were then circulated to members of the IUCN SSC-WPA Galliformes Specialist Group who had particular knowledge about a species. For some species, there were no known species experts. Specialist Group members checked information provided and supplemented it where possible. Responses varied in the level of detail returned for each species. All information is taken from BirdLife International species factsheets 2011 unless otherwise referenced. Species considered for review: Scientific name Common name Status on IUCN Red List Argusianus argus Argus pheasant Near Threatened Catreus wallichii Cheer pheasant Vulnerable Crossoptilon harmani Tibetan eared-pheasant Near Threatened Gallus sonneratii Grey junglefowl -

WPA Survey of Captive Birds 2020
Private Breeders & Zoo Collections (109) Approx 50% ringed Common name Latin name Adults Juveniles M F M F ? Breeding 12 Pairs 2020 Young Pairs Mth+ Satyr tragopan Tragopan satyra 31 32 14 19 5 Temminck's tragopan Tragopan temminckii 42 45 15 18 36 Cabot's tragopan Tragopan caboti 39 31 11 13 9 Koklass pheasant Pucrasia macrolopha 13 14 23 16 23 Himalayan monal Lophophorus impeyanus 23 25 11 7 36 Red junglefowl Gallus gallus 6 6 11 7 5 Ceylon junglefowl Gallus lafayettei 6 5 6 8 Grey junglefowl Gallus sonneratii 16 25 15 13 17 Green junglefowl Gallus varius 1 kalij pheasant - unknown subspecies Lophura leucomelana 3 1 1 White-crested kalij pheasant Lophura l. hamiltoni 5 4 2 Nepal Kalij pheasant Lophura l. leucomelana 7 7 1 4 Crawfurd's kalij pheasant Lophura l. crawfurdi Lineated kalij pheasant Lophura l. lineata 1 1 2 1 Silver pheasant - unknown subspecies Lophura nycthemera 2 3 1 True silver pheasant Lophura n. nycthemera 15 18 26 23 5 Berlioz’s silver pheasant Lophura n. berliozi 2 2 Lewis’s silver pheasant Lophura n. lewisi 5 5 3 3 Imperial pheasant Lophura imperialis 1 1 Edwards's pheasant Lophura edwardsi edwardsi 72 66 21 22 18 Vietnamese pheasant Lophura e. hatinhensis 50 45 17 20 18 Swinhoe's pheasant Lophura swinhoii 11 13 4 4 6 Salvadori's pheasant Lophura inornata 6 4 1 Malaysian crestless fireback Lophura e. erythrophthalma 7 10 4 2 2 Bornean crested fireback pheasant Lophura i. ignita/nobilis 3 4 1 1 Malaysian crestless fireback/Vieillot's Lophura i. -

Indo-Burmese Forests (PDF, 695
Indo-Burmese forests F06 F06 Threatened species HE Indo-Burmese region includes the moist tropical and Tsubtropical forests which extend from north-east India and CR EN VU Total southern China across the lowlands and isolated mountains of — 7 11 18 South-East Asia, as well as the Andaman and Nicobar islands. The forests in this region support 24 threatened bird species, 18 of which —2 35 breed nowhere else; the other six include four which also occur in the Sundaland forests (F07) and two from the South-east Chinese forests —— 11 (F03). Seven threatened species are relatively widespread within this Total — 9 15 24 region (Table 2), and 17 are confined to one of the region’s six Endemic Bird Areas and two Secondary Areas. Nine species of this Key: = breeds only in this forest region. region are Endangered, including three low-density species which = also breeds in other region(s). inhabit forested wetlands, and six restricted-range species affected by = non-breeding visitor from another region. deforestation. The Indo-Burmese forests region corresponds closely to Conservation International’s Indo-Burma Hotspot (see pp.20–21). ■ Key habitats Lowland evergreen and semi-evergreen tropical rainforest, moist deciduous forest and hill evergreen forest, and associated wetlands. ■ Altitude 0–2,400 m. ■ Countries and territories China (Yunnan, Hainan); India (Arunachal Pradesh, Assam, Meghalaya, Nagaland, Manipur, Andaman and Nicobar islands); Bhutan; Bangladesh; Myanmar; Thailand; Laos; Cambodia; Vietnam. Much of the original forest in Thailand has been cleared, and most of the remaining forests are inside protected areas such as Nam Nao National Park. -

Galliforme Guidelines 260409
Guidelines for the Re-introduction of Galliformes for Conservation Purposes Edited by the World Pheasant Association and IUCN/SSC Re-introduction Specialist Group Occasional Paper of the IUCN Species Survival Commission No. 41 IUCN Founded in 1948, IUCN (International Union for the Conservation of Nature) brings together States, government agencies and a diverse range of nongovernmental organizations in a unique world partnership: over 1000 members in all, spread across some 160 countries. As a Union, IUCN seeks to influence, encourage and assist societies throughout the world to conserve the integrity and diversity of nature and to ensure that any use of natural resources is equitable and ecologically sustainable. IUCN builds on the strengths of its members, networks and partners to enhance their capacity and to support global alliances to safeguard natural resources at local, regional and global levels. IUCN Species Survival Commission (SSC) The IUCN Species Survival Commission is a science-based network of close to 8,000 volunteer experts from almost every country of the world, all working together towards achieving the vision of, “A world that values and conserves present levels of biodiversity.” WPA-IUCN SSC Galliformes Specialist Groups The Grouse Specialist Group was established in 1993 when it developed from WPA’s long- standing grouse network. It was designed to be a global voluntary network of individuals involved in the study, conservation, and sustainable management of grouse. The Pheasant Specialist Group was established in 1991 as a voluntary self-help network of scientists, wildlife managers, conservationists, aviculturists and educators. It was particularly concerned with the plight of threatened pheasant species, and ensuring the survival of viable populations in their natural habitats whilst enhancing the quality of human life.