Yue, D.1 – Liu, X.1 – Memon, S.2 – Xi, D.1 – Deng, W.1*
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Epidemiology Bulletin
VIRGINIA EPIDEMIOLOGY BULLETIN Robert B. Stroube, M.D., M.P.H., Health Commissioner Christopher Novak, M.D., M.P.H., Editor Suzanne R. Jenkins, V.M.D., M.P.H., Acting State Epidemiologist Vickie L. O’Dell, Layout Editor January 2005 Volume 105, No. 1 Transmissible Spongiform Encephalopathies (TSEs) A little over one year ago, on Decem- of diseases known as transmissible ber 25, 2003, the US Department of Agri- spongiform encephalopathies (TSEs). This a b culture (USDA) confirmed the diagnosis article is part of the Virginia Department of bovine spongiform encephalopathy of Health’s efforts to improve surveillance (BSE, or “mad cow disease”) in a single for TSEs in Virginia to better understand 1 dairy cow in Washington state. As a re- the disease as well as to detect any conversion sult, the USDA expanded and intensified changes in disease patterns. its national testing program for BSE. From June 1, 2004, to January 2, 2005, 167,476 Overview of TSEs alpha-helices beta-sheets cattle were tested. Not a single positive TSEs are a group of relatively rare pro- 2 animal was found. gressive neurodegenerative disorders that In May 2004, the New Jersey Depart- affect both humans and animals. They are Figure 1. (a) A healthy cellular ment of Health and Senior Services distinguished by long incubation periods, prion protein contains only alpha- (NJDHSS) and the Centers for Disease characteristic spongiform changes of the helices. (b) The pathogenic isoform Control and Prevention (CDC) reported brain associated with neuronal loss, and with alpha-helical structures on a suspected cluster of Creutzfeldt- the lack of a detectable immune or inflam- converted to beta-sheets.6 Jakob disease (CJD) cases (including one matory response. -

Course Material and Quiz Answer Form (PDF)
California Association for Medical Laboratory Technology Distance Learning Program PRION DISEASES Course # DL-983 by Rebecca Rosser, MA Education & Development Consultant - Laboratory Kaiser Permanente SCPMG Regional Reference Laboratory North Hollywood, CA Approved for 1.0 CE CAMLT is approved by the California Department of Public Health as a CA CLS Accrediting Agency (#21) Level of Difficulty: Intermediate 39656 Mission Blvd. Phone: 510.792.4441 Fremont, CA 94539 FAX: 510.792.3045 Notification of Distance Learning Deadline DON’T PUT YOUR LICENSE IN JEOPARDY! This is a reminder that all the continuing education units required to renew your license/certificate must be earned no later than the expiration date printed on your license/certificate. If some of your units are made up of Distance Learning courses, please allow yourself enough time to retake the test in the event you do not pass on the first attempt. CAMLT urges you to earn your CE units early! DISTANCE LEARNING ANSWER SHEET Please Circle the one best answer COURSE NAME: PRION DISEASES COURSE # DL-983 NAME ____________________________________ LIC. # ____________________ DATE ____________ SIGNATURE (REQUIRED) ___________________________________________________________________ EMAIL_______________________________________________________________________________________ ADDRESS _____________________________________________________________________________ STREET CITY STATE/ZIP 1.0 CE – FEE: $12.00 (MEMBER) | $22.00 (NON-MEMBER) PAYMENT METHOD: [ ] CHECK OR [ ] CREDIT CARD # _____________________________________ TYPE – VISA OR MC EXP. DATE ________ | SECURITY CODE: ___ - ___ - ___ 1. a b c d 2. a b c d 3. a b c d 4. a b c d 5. a b c d 6. a b c d 7. a b c d 8. a b c d 9. a b c d 10. -
Cell Science & Stem Cell Research
Yervand E. Karapetyan, J Cell Sci Ther 2012, 3:8 http://dx.doi.org/10.4172/2157-7013.S1.022 2nd World Congress on Cell Science & Stem Cell Research November 12-14, 2012 Hilton San Antonio Airport, USA Long double stranded RNA is present in scrapie infected cells and tissues Yervand E. Karapetyan Oncological Dispensary, Artsakh Republic he nature of the infectious agent causing scrapie and other Transmissible Spongiform Encephalopathies remains an enigma Tdespite decades of research efforts. The protein-only prion hypothesis posits that abnormal conformer of a host protein is the infectious agent. Virus and virino theories include host-independent nucleic acid as the genome of the infectious agent in addition to protein component (in case of virino a host protein and in case of virus a viral protein). Viral or sub-viral nucleic acids have long been sought in scrapie to explain the existence of multiple agent strains. Many different approaches were undertaken to find such nucleic acid. Despite that no scrapie specific nucleic acid sequences have been found in infected cells or tissues. Most viruses induce synthesis of long double stranded RNA (dsRNA) during their replication in cells. Therefore the presence of long dsRNA would be an indication of viral infection in cells. J2 monoclonal antibody against long dsRNA is a great tool for easy screening of cells and tissues for the presence of suspected unknown viral infection. This antibody has not been used for testing of scrapie infected tissues. Evidence is presented here for long dsRNA in scrapie infected cells and tissues. Such dsRNA is also found in scrapie free tissue culture cells. -

Isolation of a Cdna Clone Encoding the Leader Peptide of Prion Protein and Expression of the Homologous Gene in Various Tissues
Proc. Nail. Acad. Sci. USA Vol. 83, pp. 6377-6381, September 1986 Biochemistry Isolation of a cDNA clone encoding the leader peptide of prion protein and expression of the homologous gene in various tissues (scrapie/scraple-associated fibril protein/membrane protein/prion hypothesis) NIKOLAos K. ROBAKIS*, PRITHvi R. SAWH*, GLORIA C. WOLFE*, RICHARD RUBENSTEIN*, RICHARD I. CARP*', AND MICHAEL A. INNISt *New York State Office of Mental Retardation and Developmental Disabilities, Institute for Basic Research, 1050 Forest Hill Road, Staten Island, NY 10314; and tCetus Corporation, Emeryville, CA 94608 Communicated by Fotis C. Kafatos, May 19, 1986 ABSTRACT We have isolated a hamster cDNA clone been proposed as the major structural component of the representing the coding sequences for the entire precursor of scrapie agent (14). Recently, it has been shown that NrP 27-30 prion protein (PrP) 27-30. This clone encodes a protein of 254 is encoded by a host gene, which is expressed at similar levels residues and contains an in-frame ATG codon 42 bases in normal and scrapie-infected brains. The partial nucleotide upstream from the one previously reported. Analysis of the sequence of the mRNA encoding the NrP 27-30 precursor predicted amino acid sequence suggests that the NrP precursor protein was also described (15). Immunological evidence protein contains an amino-terminal signal sequence, and a suggests that NrP 27-30 derives from the proteolytic cleavage membrane-spanigdomain in the carboxyl terminus. Cleav- of a larger precursor, NrP 33-35 (15, 16). age of the signal peptide would produce a mature protein of 232 Here we report the isolation and characterization of a amino acids. -

Vaccines and How They Work Vaccines and How They Work Vaccines and How They Work Npi Reference Guide on Vaccines and Vaccine Safety 5
VACCINES AND HOW THEY WORK VACCINES AND HOW THEY WORK VACCINES AND HOW THEY WORK NPI REFERENCE GUIDE ON VACCINES AND VACCINE SAFETY 5 GLOSSARY TERMS VACCINES AND HOW THEY WORK Acquired immunity Immunization Acute Influenza Infectious Diseases Antibody Innate immunity Antigen Lymphocytes Infectious diseases have been the bane of human existence throughout history. Association Macrophage Evidence of infection has been found in mummified remains from ancient Egypt1 Asthma Measles 2 Attenuated vaccines Mumps and elsewhere, and in the oral and written histories of all cultures. The Black Death Autoimmune disease Neisseria meningitidis of the Middles Ages,3 the decimation of the native peoples of the Americas by B cell Neonate imported disease4 and the worldwide influenza epidemic of 19184 are vivid reminders Bacteria Pathogens Booster Pentavalent vaccine that infectious diseases have profoundly shaped our world and have the potential to Bordetella pertussis Pertussis do so again. Cases Pneumococcal disease Causal association Pneumococcal polysaccharide Infectious diseases are caused by organisms that are able to exploit the human body Cell-mediated response Pneumonia so that they may grow and reproduce. In general, these organisms are tiny and can Chemokines Polysaccharide Chickenpox Polysaccharide vaccine exist in our lungs, blood or other body tissues and organs. They gain access to us Chronic Protein through the air we breathe, the food and water we ingest, or damage to our skin. Clostridium tetani Rabies Disease-causing agents (pathogens) include bacteria and viruses.5 Combination vaccine Recombinant DNA technology Conjugate vaccine Risk Bacteria are tiny and can be seen only with the aid of a microscope. -

The Clinicians' Guide to Creutzfeldt-Jakob Disease
Henry Ford Hospital Medical Journal Volume 35 Number 1 Article 17 3-1987 The Clinicians' Guide to Creutzfeldt-Jakob Disease Thomas H. Swanson Follow this and additional works at: https://scholarlycommons.henryford.com/hfhmedjournal Part of the Life Sciences Commons, Medical Specialties Commons, and the Public Health Commons Recommended Citation Swanson, Thomas H. (1987) "The Clinicians' Guide to Creutzfeldt-Jakob Disease," Henry Ford Hospital Medical Journal : Vol. 35 : No. 1 , 76-83. Available at: https://scholarlycommons.henryford.com/hfhmedjournal/vol35/iss1/17 This Article is brought to you for free and open access by Henry Ford Health System Scholarly Commons. It has been accepted for inclusion in Henry Ford Hospital Medical Journal by an authorized editor of Henry Ford Health System Scholarly Commons. Special Feature The Clinicians' Guide to Creutzfeldt-Jakob Disease Thomas H. Swanson, MD* Creutzfeldt-Jakob disease, a transmissible, rapidly progressive dementia of unknown etiology, mimics Alzheimer's disease, presents in middle life, and affects many central nervous system structures. The disease progresses in three distinct stages, culminating in death. Its occurrence is sporadic and its distribution worldwide. Pathological changes are varied, but spongy degeneration ofthe neuropil is classic. Research on scrapie, the anirrml model of Creutzfeldt-Jakob disease, has demonstrated that this unconventional, slow disease is transmitted via a small (less than 50,000 mW) particle, which is composed principally of protein. How this infectious particle, variably rmmed prion, virino, or slow virus, invokes disease or is transmitted is unclear The agent does not evoke a host immune response, nor does it appear to contain nucleic acid. -

Prions and Neuro Degenerative Diseases
African Journal of Biotechnology Vol. 10(13), pp. 2366-2374, 28 March, 2011 Available online at http://www.academicjournals.org/AJB DOI: 10.5897/AJB09.582 ISSN 1684–5315 © 2011 Academic Journals Review Prions and neuro degenerative diseases Rahul R. Nair* and Jerin K. Johnson II MTech Biotechnology, School of Biotechnology, Karunya University, Karunya Nagar, Coimbatore, Tamil NADU- 641114, India. Accepted 2 June, 2009 Prion is a disease-causing agent that is neither bacterial nor fungal nor viral and contains no genetic material. A prion is a protein that occurs normally in a harmless form. By folding into an aberrant shape, the normal prion turns into a rogue agent. It then co-opts other normal prions to become rogue prions. Prions have been held responsible for a number of degenerative brain diseases, including scrapie (a fatal disease of sheep and goats), mad cow disease, Creutzfeldt-Jacob disease, fatal familial insomnia, kuru, an unusual form of hereditary dementia known as Gertsmann-Straeussler-Scheinker disease, chronic wasting disease in deer and elk, bovine spongiform encephalopathy (BSE, commonly known as “mad-cow” disease), exotic ungulate spongiform encephalopathy, Transmissible Mink Encephalopathy (TME), and feline spongiform encephalopathy, albino tigers, pumas, and cheetahs. Currently, definite diagnosis of prion diseases is still considered to be possible only after histopathological examination of biopsied or autopsied brain material. Prion diseases do not stimulate immunity. Therefore, a vaccine that prevents them is unlikely. No treatment is available either, although research in this area is progressing. Key words: Prion, transmissible spongiform encephalopathies, diagnosis, treatment. INTRODUCTION A prion has been defined as "small proteinaceous in- holes in the brain. -

Prions: Quisling Proteins
Prions: Quisling Proteins Pat Hartigan Department of Physiology Trinity College Dublin, Ireland In the midst of the current BSE scare veterinarians What is a slow disease? are being called on for their opinion. In order to give it, The concept of slow diseases was introduced in 1954 members of the profession must first understand the eti by Sigurdsson, an Icelandic veterinarian, who observed ology of BSE and the possibility of its transmission to that diseases that run a protracted course can be divided other species. Table 2. Transmissible spongiform encephalopathies of What are transmissible spongiform animals encephalopathies? Disease Natural Host The transmissible spongiform encephalopathies Scrapie Sheep, goats (TSE) are a group of neurodegenerative diseases that constitute a subset of what were called slow diseases. Bovine spongiform TSE comprise a complex group of human and animal encephalopathy (BSE) Cattle syndromes (Tables 1 and 2) that includes genetic, infec Feline spongiform Cats, puma, tious and sporadic forms. The common characteristics encephalopathy (FSE) cheetah are spongiform (vacuolar) degeneration of neurons, re active glial changes, variable amyloid plaque formation Transmissible mink encephalopathy (TME) Mink in the C.N.S., transmissibility to experimental animals and very prolonged incubation periods (months, years), Chronic wasting disease Mule deer, elk whether the diseases are acquired naturally or experi mentally. In recent years it has emerged that Exotic ungulate Eland, oryx, encephalopathy nyala, kudu, gemsbuk. another reason for classifying them together is because the aetiology and the pathogenic mecha into two broad groups: chronic diseases and slow dis nisms that lead to clinical signs in the various eases. The course of a chronic disease tends to be diseases are related to modifications of a single irregular and is unpredictable. -

Marine-Science.Pdf
UNIVERSITY OF CALCUTTA M.Sc. Marine ScienceCourse Structure & Syllabus Semester System Effective from Academic Session 2016-17 Course Theoretical / Topics /Subjects Marks Credit Code Practical I SEMESTER MST101 Theoretical Introductory Physical Oceanography 50 4 MST102 Theoretical Fundamentals of Chemical Oceanography 50 4 MST103 Theoretical Fundamentals of Marine Geology 50 4 MST104 Theoretical Basics of Biological Oceanography 50 4 MSP105 Practical Biological & Geological Oceanography 50 4 Total 250 20 II SEMESTER MST201 Theoretical Marine Meteorology& Dynamic Oceanography 50 4 MST202 Theoretical Marine and estuarine chemistry 50 4 MST203 Theoretical Marine Geology 50 4 MST204 Theoretical Marine Ecology & Marine Pollution 50 4 MSP205 Practical Physical & Chemical Oceanography 50 4 Total 250 20 III SEMESTER MST301 Theoretical Coastal Geomorphology and Coastal Zone Management 50 4 MST302 Theoretical Marine Microbiology and Brackish Water Fisheries 50 4 management MSP303 Practical Marine Microbiology and Brackish Water Fisheries 50 4 management MSCB304 Theoretical Choice base credit system course A other than Marine Sc 50 4 MSCB305 Theoretical Choice base credit system course B other than Marine Sc 50 4 Total 250 20 IV SEMESTER MST401 Theoretical Modelling &Computational Methods in Oceanography 50 4 MSP 402 Practical Modeling & Computational Methods & in 25 2 Oceanography MST403 Theoretical Special Paper (Any One) 100 8 (i) Physical Oceanography (ii) Chemical Oceanography (iii) Biological Oceanography (iv) Geological Oceanography MSD404 Dissertation Dissertation / Review / Field work Project Submission & 75 6 Seminar Presentation and Grand Viva Voce Total 250 20 Grand Total (Semester I + II + III + IV) 1000 80 MST101: INTRODUCTORY PHYSICAL OCEANOGRAPHY Basics approaches in Physical oceanography in marine systems: Physical laws of ocean; chaos complexity & bifurcations, non-linear systems, energy paradigms: (a) ocean energy & (b) tidal energy, wave propagation in shallow water, gravity waves; Stokes’ Law and its application in marine organisms. -
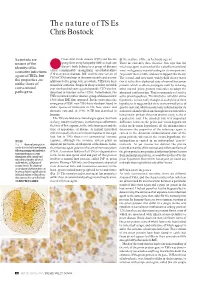
The Nature of Tses Chris Bostock
The nature of TSEs Chris Bostock Scientists are Creutzfeld–Jacob disease (CJD) and bovine ● The nature of the infectious agent unsure of the ●spongiform encephalopathy (BSE or mad cow There are currently three theories. One says that the identity of the disease) both belong to a group of diseases infectious agent is somewhat like a small conventional causative infectious called transmissible spongiform encephalopathies virus, with genetic material coding for its own proteins. (TSEs) or prion diseases. BSE and the new variant of At present there is little evidence to support this theory. agent of TSEs, but CJD (vCJD) have been in the news recently and are new The second, and now most widely held theory states the properties are additions to the group, but, as a whole, TSEs have been that it is the altered physical state of normal host prion unlike those of around for centuries. Scrapie in sheep was first recorded protein, which is able to propagate itself by inducing conventional over two hundred years ago and sporadic CJD was first other normal prion protein molecules to adopt the pathogens. described in humans in the 1920s. Nevertheless, the abnormal conformation. This is commonly referred to TSEs remained a rather obscure group of diseases until as the prion hypothesis. The third idea, called the virino 1986 when BSE first appeared. In the years since the hypothesis, is most easily thought of as a hybrid of these emergence of BSE, new TSEs have also been found in hypotheses. It suggests that there is a very small piece of exotic species of ruminants in UK zoos, exotic and genetic material, which encodes only information for its domestic cats and, in 1996, vCJD was described in own survival and replication through interaction with a humans. -

Scrapie Fact Sheet
APHIS Factsheet Veterinary Services August 2004 The scrapie agent is thought to be spread most Scrapie commonly from the ewe to her offspring and to other lambs through contact with the placenta and placen- tal fluids. Signs or effects of the disease usually Scrapie is a fatal, degenerative disease affecting the appear 2 to 5 years after the animal is infected but central nervous system of sheep and goats. It is may not appear until much later. Sheep may live 1 to among a number of diseases classified as transmissi- 6 months or longer after the onset of clinical signs, ble spongiform encephalopathies (TSE). Infected but death is inevitable. The genetics of the sheep flocks that contain a high percentage of susceptible affects their susceptibility to scrapie. animals can experience significant production losses. In the laboratory, the scrapie agent has been Over a period of several years the number of infected transmitted to hamsters, mice, rats, voles, gerbils, animals increases, and the age at onset of clinical mink, cattle, and some species of monkeys by inocu- signs decreases making these flocks economically lation. There is no scientific evidence to indicate that unviable. Female animals sold from infected flocks scrapie poses a risk to human health. There is no spread scrapie to other flocks. The presence of epidemiologic evidence that scrapie of sheep and scrapie in the United States also prevents the export goats is transmitted to humans, such as through con- of breeding stock, semen, and embryos to many tact on the farm, at slaughter plants, or butcher other countries. -

Transmissible Encephalopathies in Animals
Transmissible Encephalopathies in Animals Richard H. Kimberlin ABSTRACT their physicochemical stability and vacuolar degeneration of grey matter their apparent failure to stimulate any areas of the brain and spinal cord Scrapie in sheep and goats is the kind of protective host response to (hence the generic term, "spongiform best known of the transmissible infection. The biochemical nature of encephalopathies"). In addition brain encephalopathies of animals. The these agents is still a matter of extracts contain large numbers of combination of maternal transmission controversy but many workers con- characteristic amyloid fibrils seen by of infection and long incubation sider them to be outside the known negative stain electron microscopy (4). periods effectively maintains the range of viruses and viroids. For this These fibrils are known as scrapie- infection in flocks. A single sheep gene reason the term "unconventional associated fibrils (SAF: sometimes (Sip) controls both experimental and viruses" is often used. The terms called "prion rods") but they are an natural scrapie and the discovery of "virinos" and "prions" have entered important diagnostic feature of the allelic markers could enable the use of the literature as synonyms for the related diseases as well (5). sire selection in the control of the scrapie family of agents but this may Scrapie-associated fibrils are easily natural disease. Studies of experimen- be premature. Such terms are best purified from clinically affected brain tal rodent scrapie show that neuroin- used to identify the specific hypo- (6). They consist mainly of a mem- vasion occurs by spread of infection thetical concepts about the nature of brane glycoprotein (7) PrP, whose from visceral lymphoreticular tissues these agents that prompted their messenger RNA is present in many along nerve fibers to mid-thoracic introduction.