Transcriptome Analysis of Escherichia Coli O157:H7 During in Vivo Mimicking Conditions Michael Dwight Carruthers Iowa State University
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

12. FORMULATION of the URBAN TRANSPORT MASTER PLAN Development of the RSTP Urban Transportation Master Plan (1) Methodology
The Project on The Revision and Updating of the Strategic Transport Plan for Dhaka (RSTP) Final Report 12. FORMULATION OF THE URBAN TRANSPORT MASTER PLAN Development of the RSTP Urban Transportation Master Plan (1) Methodology The development of the RSTP Urban Transportation Master Plan adopted the following methodology (see Figure 12.1): (i) Elaborate the master plan network through a screen line analysis by comparing the network capacity and future demand. (ii) Identify necessary projects to meet future demand at the same time avoiding excessive capacity. (iii) Conducts economic evaluation of each project to give priority on projects with higher economic return. (iv) Conduct preliminary environmental assessment of every project and consider countermeasures against environmental problems, if any. (v) Make a final prioritization of all physical projects by examining their respective characteristics from different perspectives. (vi) Classify the projects into three categories, namely short-, medium- and long-term projects, by considering the financial constraints. (vii) Prepare an action plan for short-term projects together with “soft” measures. Mid-term Project Source: RSTP Study Team Figure 12.1 Development Procedure for the Master Plan 12-1 The Project on The Revision and Updating of the Strategic Transport Plan for Dhaka (RSTP) Final Report (2) Output of the Transportation Network Plan The RSTP urban transportation network plan was developed based on a review and a modification of the STP network plan. The main points of the modification or adoption of the STP network master plan are as follows: i. Harmonization with future urban structure, land-use plan and development of network plan. -

Inactivation Mechanisms of Alternative Food Processes on Escherichia Coli O157:H7
INACTIVATION MECHANISMS OF ALTERNATIVE FOOD PROCESSES ON ESCHERICHIA COLI O157:H7 DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Aaron S. Malone, M.S. ***** The Ohio State University 2009 Dissertation Committee: Approved by Professor Ahmed E. Yousef, Adviser Professor Polly D. Courtney ___________________________________ Professor Tina M. Henkin Adviser Food Science and Nutrition Professor Robert Munson ABSTRACT Application of high pressure (HP) in food processing results in a high quality and safe product with minimal impact on its nutritional and organoleptic attributes. This novel technology is currently being utilized within the food industry and much research is being conducted to optimize the technology while confirming its efficacy. Escherichia coli O157:H7 is a well studied foodborne pathogen capable of causing diarrhea, hemorrhagic colitis, and hemolytic uremic syndrome. The importance of eliminating E. coli O157:H7 from food systems, especially considering its high degree of virulence and resistance to environmental stresses, substantiates the need to understand the physiological resistance of this foodborne pathogen to emerging food preservation methods. The purpose of this study is to elucidate the physiological mechanisms of processing resistance of E. coli O157:H7. Therefore, resistance of E. coli to HP and other alternative food processing technologies, such as pulsed electric field, gamma radiation, ultraviolet radiation, antibiotics, and combination treatments involving food- grade additives, were studied. Inactivation mechanisms were investigated using molecular biology techniques including DNA microarrays and knockout mutants, and quantitative viability assessment methods. The results of this research highlighted the importance of one of the most speculated concepts in microbial inactivation mechanisms, the disruption of intracellular ii redox homeostasis. -

Effect of Nitrogen and Boron on the Yield of Wheat Cv. Kanchan
J. Bangladesh Agril. Univ. 3(2): 215-220, 2005 ISSN 1810-3030 Effect of nitrogen and boron on the yield of wheat cv. Kanchan M.M. Khan', A.K. Hasan2, M.H. Rashid2, and F. Ahrned3 Palli Karma Sahayak Foundation (PKSF), Sher-e-Banglanagar, Dhaka, 2Department of Agronomy and 3 Department of Agroforestry, Bangladesh Agricultural University, Mymensingh Abstract An experiment was carried out at the Agronomy Field Laboratory, Bangladesh Agricultural University, Mymensingh during the period from January to April 2004 to study the effect of different levels of nitrogen and boron on the yield of wheat cv. Kanchan. The treatments included four levels of nitrogen VIZ., 45, 60, 85 and 110 kg N ha-1 and four levels of boron viz., 0, 1, 2 and 3 kg B ha-1. The experiment was laid out in a Randomized Complete Block Design with three replications. The results revealed that Yield and yield contributing characters were influenced significantly by both levels of nitrogen and boron. Among the levels of nitrogen, 110 kg N ha-1 produced the highest grain (5.54 t ha-1)and straw (8.21 t ha-1 ) yields. The lowest grain (3.23 t ha-1) and straw (5.52 t ha-1) yields were observed with the application of 45 kg N ha-1. The highest grain (4.95 t ha-1)and straw (7.38 t ha 1) yields were produced With 3 kg B ha-1. The minimum grain (3.59 t ha-1)and straw (6.14 t ha-1)yields were found in the control treatment. -

Interplay Between Ompa and Rpon Regulates Flagellar Synthesis in Stenotrophomonas Maltophilia
microorganisms Article Interplay between OmpA and RpoN Regulates Flagellar Synthesis in Stenotrophomonas maltophilia Chun-Hsing Liao 1,2,†, Chia-Lun Chang 3,†, Hsin-Hui Huang 3, Yi-Tsung Lin 2,4, Li-Hua Li 5,6 and Tsuey-Ching Yang 3,* 1 Division of Infectious Disease, Far Eastern Memorial Hospital, New Taipei City 220, Taiwan; [email protected] 2 Department of Medicine, National Yang Ming Chiao Tung University, Taipei 112, Taiwan; [email protected] 3 Department of Biotechnology and Laboratory Science in Medicine, National Yang Ming Chiao Tung University, Taipei 112, Taiwan; [email protected] (C.-L.C.); [email protected] (H.-H.H.) 4 Division of Infectious Diseases, Department of Medicine, Taipei Veterans General Hospital, Taipei 112, Taiwan 5 Department of Pathology and Laboratory Medicine, Taipei Veterans General Hosiptal, Taipei 112, Taiwan; [email protected] 6 Ph.D. Program in Medical Biotechnology, Taipei Medical University, Taipei 110, Taiwan * Correspondence: [email protected] † Liao, C.-H. and Chang, C.-L. contributed equally to this work. Abstract: OmpA, which encodes outer membrane protein A (OmpA), is the most abundant transcript in Stenotrophomonas maltophilia based on transcriptome analyses. The functions of OmpA, including adhesion, biofilm formation, drug resistance, and immune response targets, have been reported in some microorganisms, but few functions are known in S. maltophilia. This study aimed to elucidate the relationship between OmpA and swimming motility in S. maltophilia. KJDOmpA, an ompA mutant, Citation: Liao, C.-H.; Chang, C.-L.; displayed compromised swimming and failure of conjugation-mediated plasmid transportation. The Huang, H.-H.; Lin, Y.-T.; Li, L.-H.; hierarchical organization of flagella synthesis genes in S. -

Connecting Bangladesh: Economic Corridor Network
Connecting Bangladesh: Economic Corridor Network Economic corridors are anchored on transport corridors, and international experience suggests that the higher the level of connectivity within and across countries, the higher the level of economic growth. In this paper, a new set of corridors is being proposed for Bangladesh—a nine-corridor comprehensive integrated multimodal economic corridor network resembling the London Tube map. This paper presents the initial results of the research undertaken as an early step of that development effort. It recommends an integrated approach to developing economic corridors in Bangladesh that would provide a strong economic foundation for the construction of world-class infrastructure that, in turn, could support the growth of local enterprises and attract foreign investment. About the Asian Development Bank COnnecTING BANGLADESH: ADB’s vision is an Asia and Pacific region free of poverty. Its mission is to help its developing member countries reduce poverty and improve the quality of life of their people. Despite the region’s many successes, it remains home to a large share of the world’s poor. ADB is committed to reducing poverty through inclusive economic growth, environmentally sustainable growth, and regional integration. ECONOMIC CORRIDOR Based in Manila, ADB is owned by 67 members, including 48 from the region. Its main instruments for helping its developing member countries are policy dialogue, loans, equity investments, guarantees, grants, NETWORK and technical assistance. Mohuiddin Alamgir -

July 2016 Volume 3 No 3
6 VOLUME 3 NO 3 JULY 2016 DHAKA CENTRAL INTERNATIONAL MEDICAL COLLEGE JOURNAL (APPROVED BY BMDC) July 2016, Vol. 3 No. 3 Contents From the Desk of Editor-in-Chief 3 Instructions for Authors 4 Editorial Novel Treatment of Diabetic Nephropathy 12 Original Articles Incidence of Malignancy in Thyroid Nodule 14 Abedin SAMA, Alam MM, Islam MS, Fakir MAY Dyslipidemia and Atherogenic Index among the 21 Young Female Doctors ofBangladesh. Khanduker S, Hoque MM, Khanduker N, Chowdhury MAA, Nazneen M A Study on Stroke in Young Patients due to Cardiac 26 Disease in a Tertiary Care Hospital in Dhaka City Mukta M, Mohammad QD, Mir AS Variation of Transverse Diameter ofDry Ossified 33 Human Atlas Vertebra of Male and Female Rahman S, Ara S, Sayeed S, Rashid S, Ferdous Z, Kashem K Study on Health Effects of Teenage Pregnancies among the Patients 36 Attending Antenatal Care Centre of Chittagong Medical College Hospital Tarafdar MA, Begum N, Das SR, Begum S, Sultana A, Rahman R, Begum R Identification ofDifferent Clinical Features and Complications of 41 Type 2 Diabetes Mellitus in Bengaladeshi Males Begum F, Shamim KM, Akter S, Hossain S, Nazma N, Afrin M, Moureen A Review Articles Female Genital Tuberculosis- A Review Article 46 Shaheed S, Mamun SMAA, Khanom M Case Reports Round Worm induced Acute Appendicitis- an Incidental 51 Finding during Colonoscopy Masum QAA, Islam MN 1 Dhaka Central International Medical College Journal Vol.13 No. 3July 2016 An Official Organ of Dhaka Central International Medical College CHIEF PATRON ADVISORS The Dhaka Central International Prof. Md. Anwarul Islam Md. -

Subunit of RNA Polymerase and the Transcriptional Regulators Rsd from Escherichia Coli and Algq from Pseudomonas Aeruginosa
Bacterial two-hybrid analysis of interactions between region 4 of the sigma(70) subunit of RNA polymerase and the transcriptional regulators Rsd from Escherichia coli and AlgQ from Pseudomonas aeruginosa. The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Dove, S. L., and A. Hochschild. 2001. “Bacterial Two-Hybrid Analysis of Interactions between Region 4 of the 70 Subunit of RNA Polymerase and the Transcriptional Regulators Rsd from Escherichia Coli and AlgQ from Pseudomonas Aeruginosa.” Journal of Bacteriology 183 (21): 6413–21. https://doi.org/10.1128/ jb.183.21.6413-6421.2001. Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:41483172 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA JOURNAL OF BACTERIOLOGY, Nov. 2001, p. 6413–6421 Vol. 183, No. 21 0021-9193/01/$04.00ϩ0 DOI: 10.1128/JB.183.21.6413–6421.2001 Copyright © 2001, American Society for Microbiology. All Rights Reserved. Bacterial Two-Hybrid Analysis of Interactions between Region 4 of the 70 Subunit of RNA Polymerase and the Transcriptional Regulators Rsd from Escherichia coli and AlgQ from Pseudomonas aeruginosa SIMON L. DOVE AND ANN HOCHSCHILD* Department of Microbiology and Molecular Genetics, Harvard Medical School, Boston, Massachusetts 02115 Received 3 May 2001/Accepted 6 August 2001 A number of transcriptional regulators mediate their effects through direct contact with the 70 subunit of Escherichia coli RNA polymerase (RNAP). -
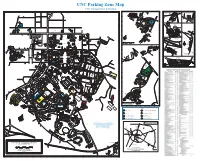
UNC Parking Zone Map UNC Transportation & Parking
UNC Parking Zone Map UNC Transportation & Parking Q R S T U V W X Y Z A B C D E F G H I J K L M N O P 26 **UNC LEASES SPACE CAROLINA . ROAD IN THESE BUILDINGS 21 21 MT HOMESTEAD NORTH LAND MGMT. PINEY OPERATIONS CTR. VD. (NC OFFICE HORACE WILLIAMS AIRPORT VD., HILL , JR. BL “RR” 41 1 1 Resident 41 CommuterRR Lot R12 UNC VD AND CHAPEL (XEROX) TE 40 MLK BL A PRINTING RIVE EXTENSION MLK BL ESTES D SERVICES TIN LUTHER KING TERST PLANT N O I AHEC T EHS HOMESTEAD ROAD MAR HANGER VD. 86) O I-40 STORAGE T R11 TH (SEE OTHER MAPS) 22 22 O 720, 725, & 730 MLK, JR. BL R1 T PHYSICAL NOR NORTH STREET ENVRNMEN HL .3 MILES TO TH. & SAFETY ESTES DRIVE 42 COMMUTER LOT T. 42 ER NC86 ELECTRICAL DISTRICENTBUTION OPERATIONS SURPLUS WA REHOUSE N1 ST GENERAL OREROOM 2 23 23 2 R1 CHAPEL HILL ES MLK JR. BOULE NORTH R1 ARKING ARD ILITI R1 / R2OVERFLOW ZONEP V VICES C R A F SHOPS GY SE EY 43 RN 43 ENERBUILDING CONSTRUCTION PRITCHARD STREET R1 NC 86 CHURCH STREET . HO , JR. BOULE ES F R1 / V STREET SER L BUILDING VICE ARD A ST ATIO GI EET N TR AIRPOR R2 S T DRIVE IN LUTHER KING BRANCH T L MAR HIL TH WEST ROSEMARY STREET EAST ROSEMARY STREET L R ACILITIES DRIVE F A NO 24 STUDRT 24 TH COLUMBI IO CHAPE R ADMINIST OFF R NO BUILDINGICE ATIVE R10 1700 N9 MLK 208 WEST 3 N10 FRANKLIN ST. -

Transcription Regulation of the Expression of the Plasmid Encoded Toxin of Enteroaggregative Escherichia Coli
TRANSCRIPTION REGULATION OF THE EXPRESSION OF THE PLASMID ENCODED TOXIN OF ENTEROAGGREGATIVE ESCHERICHIA COLI by Jacob Henry Duddy A thesis submitted to The University of Birmingham for the degree of MASTER OF PHILOSOPHY February 2013 School of Biosciences The University of Birmingham University of Birmingham Research Archive e-theses repository This unpublished thesis/dissertation is copyright of the author and/or third parties. The intellectual property rights of the author or third parties in respect of this work are as defined by The Copyright Designs and Patents Act 1988 or as modified by any successor legislation. Any use made of information contained in this thesis/dissertation must be in accordance with that legislation and must be properly acknowledged. Further distribution or reproduction in any format is prohibited without the permission of the copyright holder. Abstract The pathogenic properties of Enteroaggregative Escherichia coli strain 042 results from the synchronised expression of virulence factors, which include the Plasmid Encoded Toxin. Pet is a member of the serine protease autotransporter of the Enterobacteriaceae family and contributes to infection by cleaving α-fodrin, disrupting the actin cytoskeleton of host cells. The expression of Pet is induced by global transcription factor CRP with further enhancement by the nucleoid associated protein Fis. This study identifies the residues of RNA polymerase, Fis and CRP required for the induction of transcription, thereby clarifying the mechanism of activation employed by the transcription factors. Fis activates transcription from the Fis binding site via a direct interaction with RNA polymerase, facilitated by protein specific determinants. This interaction is dependent on the position of the Fis binding site on the DNA and it subsequent orientation on the helical face of the DNA. -

The Absence of Effect of Gid Or Mioc Transcription on the Initiation of Chromosomal Replication in Escherichia Coli (DNA Replication͞oric͞transcriptional Activation)
Proc. Natl. Acad. Sci. USA Vol. 94, pp. 12497–12502, November 1997 Genetics The absence of effect of gid or mioC transcription on the initiation of chromosomal replication in Escherichia coli (DNA replicationyoriCytranscriptional activation) DAVID B. BATES*†,ERIK BOYE‡,TSUNEAKI ASAI†§¶, AND TOKIO KOGOMA*†§i Departments of *Biology and §Cell Biology, and †Cancer Center, University of New Mexico, Albuquerque, NM 87131; and ‡Department of Cell Biology, Institute for Cancer Research, Montebello, 0310 Oslo, Norway Edited by Donald R. Helinski, University of California at San Diego, La Jolla, CA, and approved September 15, 1997 (received for review June 23, 1997) ABSTRACT Despite the widely accepted view that tran- scription of gid and mioC is required for efficient initiation of cloned oriC, we show that these transcriptions have very little effect on initiation of chromosome replication at wild-type chromosomal oriC. Furthermore, neither gid nor mioC tran- scription is required in cells deficient in the histone-like proteins Fis or IHF. However, oriC that is sufficiently im- paired for initiation by deletion of DnaA box R4 requires transcription of at least one of these genes. We conclude that transcription of mioC and especially gid is needed to activate FIG. 1. The minimal oriC and surrounding transcription. The position of the six DnaA boxes R1–R5 and M; 13-mer repeats L, M, oriC only under suboptimal conditions. We suggest that either and R; A1T-rich cluster; and binding sites for IHF and Fis proteins the rifampicin-sensitive step of initiation is some other tran- are indicated. Large arrows represent location and direction of major scription occurring from promoter(s) within oriC,orthe promoters near oriC. -

Journal of Microbiological Methods a Proteome-Wide Screen to Identify Transcription Factors Interacting with the Vibrio Cholerae
Journal of Microbiological Methods 165 (2019) 105702 Contents lists available at ScienceDirect Journal of Microbiological Methods journal homepage: www.elsevier.com/locate/jmicmeth Note A proteome-wide screen to identify transcription factors interacting with the Vibrio cholerae rpoS promoter T ⁎ ⁎ Julio C. Ayala1, Jorge A. Benitez , Anisia J. Silva Morehouse School of Medicine, Department of Microbiology, Biochemistry and Immunology, 720 Westview Dr., SW, Atlanta, GA 30310, USA ABSTRACT We describe a proteomic approach to identify transcription factors binding to a target promoter. The method's usefulness was tested by identifying proteins binding to the Vibrio cholerae rpoS promoter in response to cell density. Proteins identified in this screen included the nucleoid-associated protein Fis and the quorum sensing regulator HapR. Isolation of regulatory mutants has played a major role in increasing with agitation at 37 °C to stationary phase. Culture media were sup- our understanding of the regulation of gene expression (Shuman and plemented with ampicillin (Amp, 100 μg/mL), kanamycin (Km, 25 μg/ Silhavy, 2003). There are cases, however, in which the above approach mL), polymyxin B (PolB, 100 units/mL) or L-arabinose (0.2%) as re- can encounter significant obstacles. For instance, the methods used to quired. Chromosomal deletions of fis and/or hapR were created in strain genetically manipulate model organisms are not always equally effec- C7258ΔlacZ by allelic exchange as described previously (Wang et al., tive in less studied microorganisms; some regulatory mutations can be 2011; Wang et al., 2012; Wang et al., 2014). Briefly, genomic DNA highly pleiotropic, often be deleterious, unstable and difficult to select. -

Supplementary Material
Lacoux et al. Dynamics of transcription initiation and RNA processing Supplementary Material Dynamic insights on transcription initiation and RNA processing during bacterial adaptation Caroline Lacoux1,#, Aymeric Fouquier d’Hérouël2, Françoise Wessner- Le Bohec1, Nicolas Innocenti1,3,#, Chantal Bohn4, Sean P. Kennedy5,#, Tatiana Rochat6, Rémy A. Bonnin4,#, Pascale Serror1, Erik Aurell3,7, Philippe Bouloc4 and Francis Repoila1,* Email adresses: CL: [email protected], AFD: [email protected], FW: [email protected], NI: [email protected], CB: [email protected], SPK: [email protected], TR: [email protected], RB: [email protected], PS: [email protected], EA: [email protected], PB: [email protected], FR: [email protected] Corresponding author : [email protected] Running head: Dynamics of transcription initiation and RNA processing This file contains: Section S1: Dynamics of RNA levels in response to σE induction (Contains Figures S1 and S2). Section S2: Assignment of 5’ RNA ends. Section S3: Figure S3. Patterns of changes in TIF at σE-TSSs assigned as UNDs or PSSs in this study. Figure S4. Examples of changes in TIF at σE-independent TSSs Section S4: RNA dynamics at the bepA-yfgD operon (Contains Figures S5) References 1 Lacoux et al. Dynamics of transcription initiation and RNA processing Section S1: Dynamics of RNA levels in response to σE induction Global view of the evolving transcriptome Our experimental system consisted of a plasmid (pZE21-rpoE) that harbor the σE encoding sequence under control of the inducible promoter PLtetO-1 in E. coli K12 strain MG1655 (Fig.