New PCR Primers for Metabarcoding Environmental DNA from Freshwater
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Copyrighted Material
06_250317 part1-3.qxd 12/13/05 7:32 PM Page 15 Phylum Chordata Chordates are placed in the superphylum Deuterostomia. The possible rela- tionships of the chordates and deuterostomes to other metazoans are dis- cussed in Halanych (2004). He restricts the taxon of deuterostomes to the chordates and their proposed immediate sister group, a taxon comprising the hemichordates, echinoderms, and the wormlike Xenoturbella. The phylum Chordata has been used by most recent workers to encompass members of the subphyla Urochordata (tunicates or sea-squirts), Cephalochordata (lancelets), and Craniata (fishes, amphibians, reptiles, birds, and mammals). The Cephalochordata and Craniata form a mono- phyletic group (e.g., Cameron et al., 2000; Halanych, 2004). Much disagree- ment exists concerning the interrelationships and classification of the Chordata, and the inclusion of the urochordates as sister to the cephalochor- dates and craniates is not as broadly held as the sister-group relationship of cephalochordates and craniates (Halanych, 2004). Many excitingCOPYRIGHTED fossil finds in recent years MATERIAL reveal what the first fishes may have looked like, and these finds push the fossil record of fishes back into the early Cambrian, far further back than previously known. There is still much difference of opinion on the phylogenetic position of these new Cambrian species, and many new discoveries and changes in early fish systematics may be expected over the next decade. As noted by Halanych (2004), D.-G. (D.) Shu and collaborators have discovered fossil ascidians (e.g., Cheungkongella), cephalochordate-like yunnanozoans (Haikouella and Yunnanozoon), and jaw- less craniates (Myllokunmingia, and its junior synonym Haikouichthys) over the 15 06_250317 part1-3.qxd 12/13/05 7:32 PM Page 16 16 Fishes of the World last few years that push the origins of these three major taxa at least into the Lower Cambrian (approximately 530–540 million years ago). -

Midpacific Volume31 Issue1.Pdf
Vol. XXXI. No. 1. Lii3ky January, 1926 !A'lt 1Priqralli HMLTN CLOS ED DU 62() M5 ITED STATES AUSTRALASIA HAWAII ORIENT JAVA News Co. Gordon & Gotch Pan-Pacific Union Kelly & Walsh Javasche Boekhandel Trans-Pacific Transportation The Matson Navigation Company is palatial steamers between Honolulu and planning big things for Hawaii in many Los Angeles. The steamers visit Hilo ways. It is behind the great new Royal for the Volcano trip. The B. F. Dilling- Hawaiian Hotel at Waikiki, and is en- ham Co., Ltd., are Honolulu agents for thusing the people of Honolulu to re- the Los Angeles Steamship Company, at newed efforts to place their attractions Fort and Oueen Sts., and here may be before the people of the mainland. arranged passage direct to Los Angeles, The Company is also inducing the and beyond by rail, or you may .arrange people of Hawaii to visit California and to ship your auto or general freight. become acquainted with the people of the' scenic beaches of that state. The Mat- The Oceanic Steamship Company, son Navigation Company maintains a with head offices in San Francisco, and tourist information bureau at its main Brewer & Company as agents in Honolulu, office in the Matson Building in San maintains a fleet of swift palatial steamers Francisco, as well as in the Castle & between San Francisco, Hawaii, and Aus- Cooke Building in Honolulu, where tralia, visiting Fiji and Samoa en route. tours of the Hawaiian Islands may be This is the ideal passage to the South Seas booked. via the sunshine belt to Australasia. -

Nettastomatidae 751
click for previous page Anguilliformes: Nettastomatidae 751 NETTASTOMATIDAE Duckbill eels by D.G. Smith, National Museum of Natural History, Washington, D.C., USA iagnostic characters: Maximum size approximately 1 m, usually smaller. Body elongate, anus before Dmidlength; tail slender, attenuate, often broken and regenerated. Head slender, snout and jaws elon- gate, snout projecting a variable distance beyond tip of lower jaw. Eye well developed. Mouth large, gape extending to about rear margin of eye; no fleshy flange on upper or lower lip; some teeth exposed when mouth closed; tip of lower jaw fits into depression behind intermaxillary tooth patch. Teeth on jaws and vomer generally small, conical, multiserial, except in Hoplunnis, which has enlarged vomerine fangs. Dorsal and anal fins present, confluent with caudal fin; dorsal fin begins over or slightly behind gill opening. Pectoral fin present or absent. Scales absent. Lateral line complete. Colour: brown, lighter ventrally, without markings; dorsal and anal fins often edged in black, especially posteriorly. elongate snout and jaws Habitat, biology, and fisheries: Nettastomatids live on or near the bottom in moderate to deep water. Al- though they are occasionally taken in trawls, they have no commercial value. Similar families occurring in the area The elongate body and head and the attenuate tail distinguish the Nettastomatidae from all but a few other eels. Derichthyidae (Nessorhamphus): the jaws are elongate, but the snout is depressed and spatulate, and the posterior nostril is near the tip of the snout. Serrivomeridae: have elongate, slender jaws, but the lower jaw usually projects beyond the upper. Most spe- cies have enlarged teeth on the vomer, but these are arranged alternately in a double row, resulting in a saw-like appearance, quite different from the separated fangs present on some nettastomatids (Hoplunnis).In serrivomerids, the anterior and posterior nostrils are located close together, immediately in front of the eye. -

ASFIS ISSCAAP Fish List February 2007 Sorted on Scientific Name
ASFIS ISSCAAP Fish List Sorted on Scientific Name February 2007 Scientific name English Name French name Spanish Name Code Abalistes stellaris (Bloch & Schneider 1801) Starry triggerfish AJS Abbottina rivularis (Basilewsky 1855) Chinese false gudgeon ABB Ablabys binotatus (Peters 1855) Redskinfish ABW Ablennes hians (Valenciennes 1846) Flat needlefish Orphie plate Agujón sable BAF Aborichthys elongatus Hora 1921 ABE Abralia andamanika Goodrich 1898 BLK Abralia veranyi (Rüppell 1844) Verany's enope squid Encornet de Verany Enoploluria de Verany BLJ Abraliopsis pfefferi (Verany 1837) Pfeffer's enope squid Encornet de Pfeffer Enoploluria de Pfeffer BJF Abramis brama (Linnaeus 1758) Freshwater bream Brème d'eau douce Brema común FBM Abramis spp Freshwater breams nei Brèmes d'eau douce nca Bremas nep FBR Abramites eques (Steindachner 1878) ABQ Abudefduf luridus (Cuvier 1830) Canary damsel AUU Abudefduf saxatilis (Linnaeus 1758) Sergeant-major ABU Abyssobrotula galatheae Nielsen 1977 OAG Abyssocottus elochini Taliev 1955 AEZ Abythites lepidogenys (Smith & Radcliffe 1913) AHD Acanella spp Branched bamboo coral KQL Acanthacaris caeca (A. Milne Edwards 1881) Atlantic deep-sea lobster Langoustine arganelle Cigala de fondo NTK Acanthacaris tenuimana Bate 1888 Prickly deep-sea lobster Langoustine spinuleuse Cigala raspa NHI Acanthalburnus microlepis (De Filippi 1861) Blackbrow bleak AHL Acanthaphritis barbata (Okamura & Kishida 1963) NHT Acantharchus pomotis (Baird 1855) Mud sunfish AKP Acanthaxius caespitosa (Squires 1979) Deepwater mud lobster Langouste -

Anguilliformes, Saccopharyngiformes, and Notacanthiformes (Teleostei: Elopomorpha)
* Catalog of Type Specimens of Recent Fishes in the National Museum of Natural History, Smithsonian Institution, 6: Anguilliformes, Saccopharyngiformes, and Notacanthiformes (Teleostei: Elopomorpha) DAVID G. SMITH I SMITHSONIAN CONTRIBUTIONS TO ZOOLOGY • NUMBER 566 SERIES PUBLICATIONS OF THE SMITHSONIAN INSTITUTION Emphasis upon publication as a means of "diffusing knowledge" was expressed by the first Secretary of the Smithsonian. In his formal plan for the Institution, Joseph Henry outlined a program that included the following statement: "It is proposed to publish a series of reports, giving an account of the new discoveries in science, and of the changes made from year to year in all branches of knowledge." This theme of basic research has been adhered to through the years by thousands of titles issued in series publications under the Smithsonian imprint, commencing with Smithsonian Contributions to Knowledge in 1848 and continuing with the following active series: Smithsonian Contributions to Anthropology Smithsonian Contributions to Astrophysics Smithsonian Contributions to Botany Smithsonian Contributions to the Earth Sciences Smithsonian Contributions to the Marine Sciences Smithsonian Contributions to Paleobiology Smithsonian Contributions to Zoology Smithsonian Folklife Studies Smithsonian Studies in Air and Space Smithsonian Studies in History and Technology In these series, the Institution publishes small papers and full-scale monographs that report the research and collections of its various museums and bureaux or of professional colleagues in the world of science and scholarship. The publications are distributed by mailing lists to libraries, universities, and similar institutions throughout the world. Papers or monographs'submitted for series publication are received by the Smithsonian Institution Press, subject to its own review for format and style, only through departments of the various Smithsonian museums or bureaux, where the manuscripts are given substantive review. -

Tonga SUMA Report
BIOPHYSICALLY SPECIAL, UNIQUE MARINE AREAS OF TONGA EFFECTIVE MANAGEMENT Marine and coastal ecosystems of the Pacific Ocean provide benefits for all people in and beyond the region. To better understand and improve the effective management of these values on the ground, Pacific Island Countries are increasingly building institutional and personal capacities for Blue Planning. But there is no need to reinvent the wheel, when learning from experiences of centuries of traditional management in Pacific Island Countries. Coupled with scientific approaches these experiences can strengthen effective management of the region’s rich natural capital, if lessons learnt are shared. The MACBIO project collaborates with national and regional stakeholders towards documenting effective approaches to sustainable marine resource management and conservation. The project encourages and supports stakeholders to share tried and tested concepts and instruments more widely throughout partner countries and the Oceania region. This report outlines the process undertaken to define and describe the special, unique marine areas of Tonga. These special, unique marine areas provide an important input to decisions about, for example, permits, licences, EIAs and where to place different types of marine protected areas, locally managed marine areas and Community Conservation Areas in Tonga. For a copy of all reports and communication material please visit www.macbio-pacific.info. MARINE ECOSYSTEM MARINE SPATIAL PLANNING EFFECTIVE MANAGEMENT SERVICE VALUATION BIOPHYSICALLY SPECIAL, UNIQUE MARINE AREAS OF TONGA AUTHORS: Ceccarelli DM1, Wendt H2, Matoto AL3, Fonua E3, Fernandes L2 SUGGESTED CITATION: Ceccarelli DM, Wendt H, Matoto AL, Fonua E and Fernandes L (2017) Biophysically special, unique marine areas of Tonga. MACBIO (GIZ, IUCN, SPREP), Suva. -
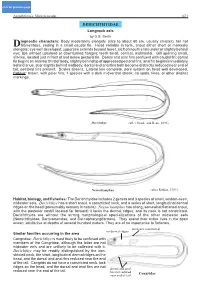
DERICHTHYIDAE Longneck Eels by D.G
click for previous page Anguilliformes: Muraenesocidae 1671 DERICHTHYIDAE Longneck eels by D.G. Smith iagnostic characters: Body moderately elongate (size to about 60 cm, usually smaller), tail not Dfilamentous, ending in a small caudal fin. Head variable in form, snout either short or markedly elongate; eye well developed; upper jaw extends beyond lower, cleft of mouth ends under or slightly behind eye; lips without upturned or downturned flanges; teeth small, conical, multiserial. Gill opening small, slit-like, located just in front of and below pectoral fin. Dorsal and anal fins confluent with caudal fin; dorsal fin begins on anterior third of body, slightly behind tip of appressed pectoral fins; anal fin begins immediately behind anus, at or slightly behind midbody; dorsal and anal fins both become distinctly reduced near end of tail; pectoral fins present. Scales absent. Lateral line complete, pore system on head well developed. Colour: brown, with paler fins; 1 species with a dark midventral streak; no spots, lines, or other distinct markings. Derichthys (after Goode and Bean, 1896) Nessorhamphus (after Robins, 1989) Habitat, biology, and fisheries: The Derichthyidae includes 2 genera and 3 species of small, seldom-seen, midwater eels. Derichthys has a short snout, a constricted neck, and a series of short, longitudinal dermal ridges on the head (presumably sensory in nature). Nessorhamphus has a long, somewhat flattened snout, with the posterior nostril located far forward; it lacks the dermal ridges, and its neck is not constricted. Derichthyids are without the strong morphological specializations of the other midwater eels (Nemichthyidae, Serrivomeridae, and Saccopharyngiformes). They spend their entire lives in the open ocean; adults live at depths of several hundred meters. -

Record of the Rare Caribbean Mud Eel
ACTA AMAZONICA http://dx.doi.org/10.1590/1809-4392201802883 ORIGINAL ARTICLE Record of the rare Caribbean mud eel, Pythonichthys sanguineus (Heterenchelyidae, Anguilliformes), in the region of the Amazon Reef Matheus Marcos ROTUNDO1, Leonardo MACHADO2, Claudio OLIVEIRA2, Wagner César Rosa dos SANTOS3, Alexandre Pires MARCENIUK2,3,* 1 Universidade Santa Cecília, Acervo Zoológico, Santos, SP, Brasil. 2 Universidade Estadual Paulista, Departamento de Morfologia, Botucatu, SP, Brasil. 3 Centro de Pesquisa e Gestão de Recursos Pesqueiros do Litoral Norte (CEPNOR), Belém, Pará, Brasil. * Corresponding author: [email protected] ABSTRACT As they spend most of their time buried in the substrate and are not a fishery resource, heterenchelyids are seldom seen. These eels are characterized by their greatly reduced eyes, which are covered by semi-transparent skin, the absence of a pectoral finor lateral line, and no pores on the head or body. Pythonichthys sanguineus is a particularly poorly-known species, with only eight scientific records from Cuba, Puerto Rico, Colombia, Venezuela, Guyana, and Suriname. The present study is based on six adult specimens of P. sanguineus captured by vessels of the shrimp trawling fleet along the northern Brazilian coast, in the vicinity of the Amazon Reef. We provide meristic, morphometric and DNA barcoding data. These findings provide insights into the distribution of the species off the northern Brazilian coast and contribute to the discussion about the southern limit of the Greater Caribbean fauna. KEYWORDS: fisheries, conservation, Brazilian northern coast, new record, systematics Registro da rara enguia de lama do Caribe, Pythonichthys sanguineus (Heterenchelyidae, Anguilliformes) na região dos Recifes da Amazônia RESUMO Heterenchelídeos passam a maior parte do tempo enterrados no substrato e raramente são vistos, não representando recursos pesqueiros importantes. -

Trophic Structure and Food Resources of Epipelagic and Mesopelagic Fishes in the North Pacific Subtropical Gyre Ecosystem Inferred from Nitrogen Isotopic Compositions
LIMNOLOGY and Limnol. Oceanogr. 60, 2015, 1156–1171 OCEANOGRAPHY VC 2015 Association for the Sciences of Limnology and Oceanography doi: 10.1002/lno.10085 Trophic structure and food resources of epipelagic and mesopelagic fishes in the North Pacific Subtropical Gyre ecosystem inferred from nitrogen isotopic compositions C. Anela Choy,*1,2 Brian N. Popp,3 Cecelia C. S. Hannides,1,3 Jeffrey C. Drazen1 1Department of Oceanography, University of Hawaii at Manoa, Honolulu, Hawaii 2Present address: Monterey Bay Aquarium Research Institute, Moss Landing, California 3Department of Geology & Geophysics, University of Hawaii at Manoa, Honolulu, Hawaii Abstract We used bulk tissue d13C and d15N values and d15N values of individual amino acids (AA) to characterize the trophic structure of a pelagic fish assemblage from the North Pacific Subtropical Gyre (NPSG) ecosystem. We focus on energy flow between fishes inhabiting distinct epipelagic, mesopelagic, and upper bathypelagic habitats and on predatory fish foraging across and within these depth habitats. Trophic positions (TPs) esti- 15 mated from a combination of trophic and source AA d N values (TPTr-Src) spanned a narrow range of 0.7 TP for 10 species of large fishes, including tunas, billfishes, and gempylids (TPTr-Src 4.3-5.0). Similarly, 13 species 15 of small micronekton fishes encompassed a range of 1.2 TP (TPTr-Src 2.6-3.8). The d N values of three source 15 AAs were found to increase with increasing depth of capture across the 13 micronekton fish species (d NPhe 15 15 range 5 6.6&; d NGly range 5 13.4&; d NSer range 5 13.6&), indicating that some species from epipelagic, mesopelagic, and upper bathypelagic communities access distinct food resources, such as suspended particles. -

Fishes of the World
Fishes of the World Fishes of the World Fifth Edition Joseph S. Nelson Terry C. Grande Mark V. H. Wilson Cover image: Mark V. H. Wilson Cover design: Wiley This book is printed on acid-free paper. Copyright © 2016 by John Wiley & Sons, Inc. All rights reserved. Published by John Wiley & Sons, Inc., Hoboken, New Jersey. Published simultaneously in Canada. No part of this publication may be reproduced, stored in a retrieval system, or transmitted in any form or by any means, electronic, mechanical, photocopying, recording, scanning, or otherwise, except as permitted under Section 107 or 108 of the 1976 United States Copyright Act, without either the prior written permission of the Publisher, or authorization through payment of the appropriate per-copy fee to the Copyright Clearance Center, 222 Rosewood Drive, Danvers, MA 01923, (978) 750-8400, fax (978) 646-8600, or on the web at www.copyright.com. Requests to the Publisher for permission should be addressed to the Permissions Department, John Wiley & Sons, Inc., 111 River Street, Hoboken, NJ 07030, (201) 748-6011, fax (201) 748-6008, or online at www.wiley.com/go/permissions. Limit of Liability/Disclaimer of Warranty: While the publisher and author have used their best efforts in preparing this book, they make no representations or warranties with the respect to the accuracy or completeness of the contents of this book and specifically disclaim any implied warranties of merchantability or fitness for a particular purpose. No warranty may be createdor extended by sales representatives or written sales materials. The advice and strategies contained herein may not be suitable for your situation. -

魚 類 学 雑 誌 総 説 ・Review 48(2):75-91
魚 類 学 雑 誌 総 説 ・Review 48(2):75-91 カ ラ イ ワ シ類(Elopomorpha)を 中 心 と した 下 位 真 骨 類 の 系 統 井 上 潤 上 宮 正 樹2 1〒164 -8639中 野 区 南 台1-15-1東 京 大 学 海 洋 研 究 所 2〒260 -8682千 葉 市 中 央 区 青 葉 町955-2千 葉 県 立 中 央 博 物 館 (2001年3月15日 受 付;2001年5月16日 改 訂;2001年5月21日 受 理) キ ー ワ ー ド:下 位 真 骨 類 の分 類 体 系,カ ラ イ ワ シ類 の 単 系 統 性,レ プ トケ フ ァ ル ス 幼 生,ミ トゲ ノ ム Jun G. Inoue* and Masaki Miya. 2001. Phylogeny of the basal teleosts, with spe- 魚 類 学 雑 談 cial reference to the Elopomorpha. Japan. J. Ichthyol., 48 (2): 75-91. Japanese Journal of Ichthyology Abstract Higher-level relationships of the basal teleosts are reviewed, with par- TheIchthyological (c) Societyof Japan 2001 ticular reference to the monophyly and interrelationships of the Elopomorpha. Since Greenwood et al. established in 1966 the Elopomorpha on the basis of eight putative synapomorphies, many authors have discussed validity of their mono- phyly through comparative observations of osteology, ontogeny, microstructure of spermatozoa, and physiology. Such circumstantial evidence have remained contro- versial, because none of them have reconstructed their phylogeny using character matrices derived from both morphological and molecular data. The same is true of long-standing controversy over phylogenetic positions of the Elopomorpha among major basal teleostean lineages, many authors simply demonstrating their "views" in the form of cladograms with putative synapomorphies until quite recently. -

Zootaxa, the Deep-Sea Anguilliformes and Saccopharyngiformes
Zootaxa 2234: 1–20 (2009) ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ Article ZOOTAXA Copyright © 2009 · Magnolia Press ISSN 1175-5334 (online edition) The deep-sea Anguilliformes and Saccopharyngiformes (Teleostei: Elopomorpha) collected on the Brazilian continental slope, between 11o and 23o S MARCELO R. S. MELO1,4, GUSTAVO W. A. NUNAN2, ADRIANA C. BRAGA3 & PAULO A. S. COSTA3 1 331 Funchess Hall, Auburn University, Auburn, AL 36849 USA. E-mail: [email protected] 2 Dept. de Vertebrados, Museu Nacional/UFRJ, Quinta da Boa Vista, Rio de Janeiro, RJ 20940–040 Brazil 3 Laboratório de Dinâmica de Populações, Universidade Federal do Estado do Rio de Janeiro – UNIRIO, Av. Pasteur, 458, ECB sala 410, Urca, Rio de Janeiro, RJ 22290-240 Brazil, Email: [email protected]; [email protected]. 4 Corresponding author Abstract A review of the deep-sea anguilliform and saccopharyngiform eels collected by the French R/Vs Marion Dufresne (1987), Thalassa (1999, 2000), and the Brazilian Astro Garoupa (2003) revealed a great diversity of these groups on the Brazilian continental slope (11–23o S, 19–40o W), in the depth range of 233 to 3450 m. Of the 33 species collected, 13 (39.4%) are being reported for the first time in the western South Atlantic. New taxa are represented by one species recently described and few other are probably undescribed. The most species-rich family in the area was Synaphobranchidae (11 species), followed by Congridae (9), Nettastomatidae (5), Nemichthyidae (3), Serrivomeridae (2), Colocongridae (1), Cyematidae (1), and Eurypharyngidae (1). Regarding the vertical species distribution, a gradual transition of species was observed, without any clear break along the slope.