The Guanine and Cytosine Content of Genomic DNA and Bacterial Evolution
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Guardian of the Development of Diabetic Retinopathy
Diabetes Volume 67, April 2018 745 Sirt1: A Guardian of the Development of Diabetic Retinopathy Manish Mishra, Arul J. Duraisamy, and Renu A. Kowluru Diabetes 2018;67:745–754 | https://doi.org/10.2337/db17-0996 Diabetic retinopathy is a multifactorial disease, and the molecular mechanism of the development of diabetic reti- exact mechanism of its pathogenesis remains obscure. nopathy remains to be established. Sirtuin 1 (Sirt1), a multifunctional deacetylase, is impli- Sirtuin 1 (Sirt1), a member of the silent information cated in the regulation of many cellular functions and in regulator 2 family, is a class III histone deacetylase that gene transcription, and retinal Sirt1 is inhibited in di- interacts with target proteins and regulates many cellular abetes. Our aim was to determine the role of Sirt1 in the functions including cell proliferation, apoptosis, and inflam- development of diabetic retinopathy and to elucidate the matory responses (6–8). Sirt1 is mainly a nuclear protein, Sirt1 molecular mechanism of its downregulation. Using - and its activity depends on cellular NAD availability (9). It is overexpressing mice that were diabetic for 8 months, Sirt1 expressed throughout the retina, and upregulation of COMPLICATIONS structural, functional, and metabolic abnormalities were protects against various ocular diseases including retinal investigated in vascular and neuronal retina. The role of degeneration, cataract, and optic neuritis (10). Our previous epigenetics in Sirt1 transcriptional suppression was inves- work has shown that Sirt1 expression and activity are de- tigated in retinal microvessels. Compared with diabetic wild-type mice, retinal vasculature from diabetic Sirt1 mice creased in the retina and its capillary cells in diabetes (11). -

The Transcriptional Landscape of a Rewritten Bacterial Genome Reveals Control Elements and Genome Design Principles ✉ ✉ Mariëlle J
ARTICLE https://doi.org/10.1038/s41467-021-23362-y OPEN The transcriptional landscape of a rewritten bacterial genome reveals control elements and genome design principles ✉ ✉ Mariëlle J. F. M. van Kooten 1 , Clio A. Scheidegger1, Matthias Christen1 & Beat Christen 1 Sequence rewriting enables low-cost genome synthesis and the design of biological systems with orthogonal genetic codes. The error-free, robust rewriting of nucleotide sequences can 1234567890():,; be achieved with a complete annotation of gene regulatory elements. Here, we compare transcription in Caulobacter crescentus to transcription from plasmid-borne segments of the synthesized genome of C. ethensis 2.0. This rewritten derivative contains an extensive amount of supposedly neutral mutations, including 123’562 synonymous codon changes. The tran- scriptional landscape refines 60 promoter annotations, exposes 18 termination elements and links extensive transcription throughout the synthesized genome to the unintentional intro- duction of sigma factor binding motifs. We reveal translational regulation for 20 CDS and uncover an essential translational regulatory element for the expression of ribosomal protein RplS. The annotation of gene regulatory elements allowed us to formulate design principles that improve design schemes for synthesized DNA, en route to a bright future of iteration- free programming of biological systems. 1 Institute of Molecular Systems Biology, Department of Biology, Eidgenössische Technische Hochschule Zürich, Zürich, Switzerland. ✉ email: [email protected]; [email protected] NATURE COMMUNICATIONS | (2021) 12:3053 | https://doi.org/10.1038/s41467-021-23362-y | www.nature.com/naturecommunications 1 ARTICLE NATURE COMMUNICATIONS | https://doi.org/10.1038/s41467-021-23362-y e can program biological systems with DNA that is In previous work, we synthesized and assembled the rewritten Wbased on native nucleotide sequences. -

Cytosine-Rich
Proc. Natl. Acad. Sci. USA Vol. 93, pp. 12116-12121, October 1996 Chemistry Inter-strand C-H 0 hydrogen bonds stabilizing four-stranded intercalated molecules: Stereoelectronic effects of 04' in cytosine-rich DNA (base-ribose stacking/sugar pucker/x-ray crystallography) IMRE BERGERt, MARTIN EGLIt, AND ALEXANDER RICHt tDepartment of Biology, Massachusetts Institute of Technology, Cambridge, MA 02139; and tDepartment of Molecular Pharmacology and Biological Chemistry, Northwestern University Medical School, 303 East Chicago Avenue, Chicago, IL 60611-3008 Contributed by Alexander Rich, August 19, 1996 ABSTRACT DNA fragments with stretches of cytosine matic cytosine ring systems from intercalated duplexes (Fig. 1A). residues can fold into four-stranded structures in which two Second, unusually close intermolecular contacts between sugar- parallel duplexes, held together by hemiprotonated phosphate backbones in the narrow grooves are observed, with cytosine-cytosine+ (C C+) base pairs, intercalate into each inter-strand phosphorus-phosphorus distances as close as 5.9 A other with opposite polarity. The structural details of this (5), presumably resulting in unfavorable electrostatic repulsion if intercalated DNA quadruplex have been assessed by solution not shielded by cations or bridging water molecules. NMR and single crystal x-ray diffraction studies of cytosine- The close contacts between pairs of antiparallel sugar- rich sequences, including those present in metazoan telo- phosphate backbones from the two interdigitated duplexes are meres. A conserved feature of these structures is the absence a unique characteristic of four-stranded intercalated DNA. of stabilizing stacking interactions between the aromatic ring Indeed, the unusually strong nuclear overhauser effect signals systems of adjacent C-C+ base pairs from intercalated du- between inter-strand sugar Hi' protons and Hi' and H4' plexes. -

REDUCTION of PURINE CONTENT in COMMONLY CONSUMED MEAT PRODUCTS THROUGH RINSING and COOKING by Anna Ellington (Under the Directio
REDUCTION OF PURINE CONTENT IN COMMONLY CONSUMED MEAT PRODUCTS THROUGH RINSING AND COOKING by Anna Ellington (Under the direction of Yen-Con Hung) Abstract The commonly consumed meat products ground beef, ground turkey, and bacon were analyzed for purine content before and after a rinsing treatment. The rinsing treatment involved rinsing the meat samples using a wrist shaker in 5:1 ratio water: sample for 2 or 5 minutes then draining or centrifuging to remove water. The total purine content of 25% fat ground beef significantly decreased (p<0.05) from 8.58 mg/g protein to a range of 5.17-7.26 mg/g protein after rinsing treatments. After rinsing and cooking an even greater decrease was seen ranging from 4.59-6.32 mg/g protein. The total purine content of 7% fat ground beef significantly decreased from 7.80 mg/g protein to a range of 5.07-5.59 mg/g protein after rinsing treatments. A greater reduction was seen after rinsing and cooking in the range of 4.38-5.52 mg/g protein. Ground turkey samples showed no significant changes after rinsing, but significant decreases were seen after rinsing and cooking. Bacon samples showed significant decreases from 6.06 mg/g protein to 4.72 and 4.49 after 2 and 5 minute rinsing and to 4.53 and 4.68 mg/g protein after 2 and 5 minute rinsing and cooking. Overall, this study showed that rinsing foods in water effectively reduces total purine content and subsequent cooking after rinsing results in an even greater reduction of total purine content. -

Chapter 23 Nucleic Acids
7-9/99 Neuman Chapter 23 Chapter 23 Nucleic Acids from Organic Chemistry by Robert C. Neuman, Jr. Professor of Chemistry, emeritus University of California, Riverside [email protected] <http://web.chem.ucsb.edu/~neuman/orgchembyneuman/> Chapter Outline of the Book ************************************************************************************** I. Foundations 1. Organic Molecules and Chemical Bonding 2. Alkanes and Cycloalkanes 3. Haloalkanes, Alcohols, Ethers, and Amines 4. Stereochemistry 5. Organic Spectrometry II. Reactions, Mechanisms, Multiple Bonds 6. Organic Reactions *(Not yet Posted) 7. Reactions of Haloalkanes, Alcohols, and Amines. Nucleophilic Substitution 8. Alkenes and Alkynes 9. Formation of Alkenes and Alkynes. Elimination Reactions 10. Alkenes and Alkynes. Addition Reactions 11. Free Radical Addition and Substitution Reactions III. Conjugation, Electronic Effects, Carbonyl Groups 12. Conjugated and Aromatic Molecules 13. Carbonyl Compounds. Ketones, Aldehydes, and Carboxylic Acids 14. Substituent Effects 15. Carbonyl Compounds. Esters, Amides, and Related Molecules IV. Carbonyl and Pericyclic Reactions and Mechanisms 16. Carbonyl Compounds. Addition and Substitution Reactions 17. Oxidation and Reduction Reactions 18. Reactions of Enolate Ions and Enols 19. Cyclization and Pericyclic Reactions *(Not yet Posted) V. Bioorganic Compounds 20. Carbohydrates 21. Lipids 22. Peptides, Proteins, and α−Amino Acids 23. Nucleic Acids ************************************************************************************** -

Human Hypoxanthine (Guanine) Phosphoribosyltransferase: An
Proc. NatL Acad. Sci. USA Vol. 80, pp. 870-873, Febriary 1983 Medical Sciences Human hypoxanthine (guanine) phosphoribosyltransferase: An amino acid substitution in a mutant form of the enzyme isolated from a patient with gout (reverse-phase HPLC/peptide mapping/mutant enzyme) JAMES M. WILSON*t, GEORGE E. TARRt, AND WILLIAM N. KELLEY*t Departments of *Internal Medicine and tBiological Chemistry, University of Michigan Medical School, Ann Arbor, Michigan 48109 Communicated by James B. Wyngaarden, November 3, 1982 ABSTRACT We have investigated the molecular basis for a tration ofenzyme protein. in both erythrocytes (3) and lympho- deficiency ofthe enzyme hypoxanthine (guanine) phosphoribosyl- blasts (4); (ii) a normal Vm., a normal Km for 5-phosphoribosyl- transferase (HPRT; IMP:pyrophosphate-phosphoribosyltransfer- 1-pyrophosphate, and a 5-fold increased Km for hypoxanthine ase, EC 2.4.2.8) in a patient with a severe form of gout. We re-. (unpublished data); (iii) a normal isoelectric point (3, 4) and ported in previous studies the isolation of a unique structural migration during nondenaturing polyacrylamide gel electro- variant of HPRT from this patient's erythrocytes and cultured phoresis (4); and (iv) -an apparently smaller subunit molecular lymphoblasts. This enzyme variant, which is called HPRTOnd0., weight as evidenced by an increased mobility during Na- is characterized by a decreased concentration of HPRT protein DodSO4/polyacrylamide gel electrophoresis (3, 4). in erythrocytes and lymphoblasts, a normal Vm.., a 5-fold in- Our study ofthe tryptic peptides and amino acid composition creased Km for hypoxanthine, a normal isoelectric point, and an of apparently smaller subunit molecular weight. Comparative pep- HPRTLondon revealed a single amino acid substitution (Ser tide mapping-experiments revealed a single abnormal tryptic pep- Leu) at position 109. -

Horizontal Gene Flow Into Geobacillus Is Constrained by the Chromosomal Organization of Growth and Sporulation
bioRxiv preprint doi: https://doi.org/10.1101/381442; this version posted August 2, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Horizontal gene flow into Geobacillus is constrained by the chromosomal organization of growth and sporulation Alexander Esin1,2, Tom Ellis3,4, Tobias Warnecke1,2* 1Molecular Systems Group, Medical Research Council London Institute of Medical Sciences, London, United Kingdom 2Institute of Clinical Sciences, Faculty of Medicine, Imperial College London, London, United Kingdom 3Imperial College Centre for Synthetic Biology, Imperial College London, London, United Kingdom 4Department of Bioengineering, Imperial College London, London, United Kingdom *corresponding author ([email protected]) 1 bioRxiv preprint doi: https://doi.org/10.1101/381442; this version posted August 2, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Abstract Horizontal gene transfer (HGT) in bacteria occurs in the context of adaptive genome architecture. As a consequence, some chromosomal neighbourhoods are likely more permissive to HGT than others. Here, we investigate the chromosomal topology of horizontal gene flow into a clade of Bacillaceae that includes Geobacillus spp. Reconstructing HGT patterns using a phylogenetic approach coupled to model-based reconciliation, we discover three large contiguous chromosomal zones of HGT enrichment. -

Section 4. Guidance Document on Horizontal Gene Transfer Between Bacteria
306 - PART 2. DOCUMENTS ON MICRO-ORGANISMS Section 4. Guidance document on horizontal gene transfer between bacteria 1. Introduction Horizontal gene transfer (HGT) 1 refers to the stable transfer of genetic material from one organism to another without reproduction. The significance of horizontal gene transfer was first recognised when evidence was found for ‘infectious heredity’ of multiple antibiotic resistance to pathogens (Watanabe, 1963). The assumed importance of HGT has changed several times (Doolittle et al., 2003) but there is general agreement now that HGT is a major, if not the dominant, force in bacterial evolution. Massive gene exchanges in completely sequenced genomes were discovered by deviant composition, anomalous phylogenetic distribution, great similarity of genes from distantly related species, and incongruent phylogenetic trees (Ochman et al., 2000; Koonin et al., 2001; Jain et al., 2002; Doolittle et al., 2003; Kurland et al., 2003; Philippe and Douady, 2003). There is also much evidence now for HGT by mobile genetic elements (MGEs) being an ongoing process that plays a primary role in the ecological adaptation of prokaryotes. Well documented is the example of the dissemination of antibiotic resistance genes by HGT that allowed bacterial populations to rapidly adapt to a strong selective pressure by agronomically and medically used antibiotics (Tschäpe, 1994; Witte, 1998; Mazel and Davies, 1999). MGEs shape bacterial genomes, promote intra-species variability and distribute genes between distantly related bacterial genera. Horizontal gene transfer (HGT) between bacteria is driven by three major processes: transformation (the uptake of free DNA), transduction (gene transfer mediated by bacteriophages) and conjugation (gene transfer by means of plasmids or conjugative and integrated elements). -

Motoo Kimura (1924-1994)
Motoo Kimura (1924-1994) OR decades the field of mathematical population prize-winners. When our textbook (CROW and KIMURA F genetics and evolutionary theory was dominated 1970) was published, he used his royalties to build a by the three pioneers,J. B. S. HALDANE,R. A. FISHER, tiny greenhouse attached to his home. Every Sunday and SEM'A1.L. WRIGHT.M'ith WRIGI-IT'Sdeath (CROW was orchid day. He used his artistic talent to paint pic- 1988), and for some time before, the leadingsuccessor tures of his favorite flowers, usually on chinaware. to this great heritage was MOTOO KIMURA.Although From age 17 to 19 KIMURA was in high school, where best known for his daring neutral theory of molecular a friendly and scientifically literate teacher encouraged evolution, a concept of great interest andequally great his study of chromosome morphology, and hebecame controversy, he is admired by populationgeneticists a plant cytogeneticist. At that time, cytogenetics was even more forhis deep contributions to the mathemati- very popular in Japan,and he joined thearmy of chro- cal theory. mosome watchers. During this period he was also fasci- MOTOO KIMURA was born November 13,1924 in Oka- nated by a physics course. HIDEKIYUKAWA, later to win zaki, Japan. He diedNovember 13, 1994, on theseventi- the Nobel Prize for predicting the meson, became his eth anniversary of his birth. For some timehe had been scientific hero, and KIMURA began to take an interest a victim of amyotrophic lateral sclerosis and was pro- in mathematics as the language of science. gressively weakening. Nevertheless, his death was acci- Japan was then in themidst of World War 11, and the dental. -

Detecting Selection on Noncoding Nucleotide Variation: Methods and Applications
University of Pennsylvania ScholarlyCommons Publicly Accessible Penn Dissertations 2015 Detecting Selection on Noncoding Nucleotide Variation: Methods and Applications Yang Ding University of Pennsylvania, [email protected] Follow this and additional works at: https://repository.upenn.edu/edissertations Part of the Bioinformatics Commons, Biology Commons, and the Evolution Commons Recommended Citation Ding, Yang, "Detecting Selection on Noncoding Nucleotide Variation: Methods and Applications" (2015). Publicly Accessible Penn Dissertations. 1687. https://repository.upenn.edu/edissertations/1687 This paper is posted at ScholarlyCommons. https://repository.upenn.edu/edissertations/1687 For more information, please contact [email protected]. Detecting Selection on Noncoding Nucleotide Variation: Methods and Applications Abstract There has been a long tradition in molecular evolution to study selective pressures operating at the amino-acid level. But protein-coding variation is not the only level on which molecular adaptations occur, and it is not clear what roles non-coding variation has played in evolutionary history, since they have not yet been systematically explored. In this dissertation I systematically explore several aspects of selective pressures of noncoding nucleotide variation: The first project (Chapter 2) describes research on the determinants of eukaryotic translation dynamics, which include selection on non-coding aspects of DNA variation. Deep sequencing of ribosome-protected mRNA fragments and polysome gradients in various eukaryotic organisms have revealed an intriguing pattern: shorter mRNAs tend to have a greater overall density of ribosomes than longer mRNAs. There is debate about the cause of this trend. To resolve this open question, I systematically analysed 5’ mRNA structure and codon usage patterns in short versus long genes across 100 sequenced eukaryotic genomes. -

Pdfs/0551.Pdf
1 Structures of the Molecular Components in DNA and RNA with Bond Lengths Interpreted as Sums of Atomic Covalent Radii Raji Heyrovská* Institute of Biophysics of the Academy of Sciences of the Czech Republic, Královopolská 135, 61265 Brno, Czech Republic. *E-mail: [email protected] Abstract: The author has found recently that the lengths of chemical bonds are sums of the covalent and or ionic radii of the relevant atoms constituting the bonds, whether they are completely or partially covalent or ionic. This finding has been tested here for the skeletal bond lengths in the molecular constituents of nucleic acids, adenine, thymine, guanine, cytosine, uracil, ribose, deoxyribose and phosphoric acid. On collecting the existing data and comparing them graphically with the sums of the appropriate covalent radii of C, N, O, H and P, it is found that there is a linear dependence with unit slope and zero intercept. This shows that the bond lengths in the above molecules can be interpreted as sums of the relevant atomic covalent radii. Based on this, the author has presented here (for the first time) the atomic structures of the above molecules and of DNA and RNA nucleotides with Watson-Crick base pairs, which satisfy the known dimensions. INTRODUCTION Following the exciting discovery [1] of the molecular structure of nucleic acids, the question of the skeletal bond lengths in the molecules constituting DNA and RNA has continued to be of interest. The author was enthused to undertake this work by the recent findings [2a, b] that the length of the chemical bond between any two atoms or ions is, in general, the sum of the radii of 2 the atoms and or ions constituting the bond. -
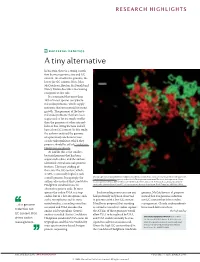
Bacterial Genetics a Tiny Alternative
RESEARCH HIGHLIGHTS BACTERIAL GENETICS A tiny alternative In bacteria, there is a strong correla- tion between genome size and GC content: the smaller the genome, the lower the GC content. Now, John McCutcheon, Bradon McDonald and Nancy Moran describe a fascinating exception to this rule. It is estimated that more than 10% of insect species carry bacte- rial endosymbionts, which supply nutrients that are essential for insect growth. The genomes of the bacte- rial endosymbionts that have been sequenced so far are much smaller than the genomes of other intracel- lular or free-living bacteria and all have a low GC content. In this study, the authors analysed the genome of a previously uncharacterized cicada endosymbiont, which they propose should be called Candidatus Hodgkinia cicadicola. At 144 kb, this is the smallest bacterial genome that has been sequenced to date, and the authors identified several unusual genomic features. The most striking of these was the GC content, which, at 58%, is unusually high for such Micrograph showing Candidatus Hodgkinia cicadicola (red) in close association with another endosymbiont, a small genome. Intriguingly, the Candidatus Sulcia muelleri (green) in the cicada Diceroprocta semicincta. The scale bar represents 10 µm. authors also noticed that Candidatus Image reproduced from McCutcheon, J. P., McDonald, B. R. & Moran, N. P. Origin of an alternative genetic Hodgkinia cicadicola uses an code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 5, e1000565 (2009). alternative genetic code. In most species the codon UGA is a stop Such recoding events are rare and genome, McCutcheon et al.