GEDEVO: an Evolutionary Graph Edit Distance Algorithm for Biological Network Alignment
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Assigning Folds to the Proteins Encoded by the Genome of Mycoplasma Genitalium (Protein Fold Recognition͞computer Analysis of Genome Sequences)
Proc. Natl. Acad. Sci. USA Vol. 94, pp. 11929–11934, October 1997 Biophysics Assigning folds to the proteins encoded by the genome of Mycoplasma genitalium (protein fold recognitionycomputer analysis of genome sequences) DANIEL FISCHER* AND DAVID EISENBERG University of California, Los Angeles–Department of Energy Laboratory of Structural Biology and Molecular Medicine, Molecular Biology Institute, University of California, Los Angeles, Box 951570, Los Angeles, CA 90095-1570 Contributed by David Eisenberg, August 8, 1997 ABSTRACT A crucial step in exploiting the information genitalium (MG) (10), as a test of the capabilities of our inherent in genome sequences is to assign to each protein automatic fold recognition server and as a case study to sequence its three-dimensional fold and biological function. identify the difficulties facing automated fold assignment. Here we describe fold assignment for the proteins encoded by the small genome of Mycoplasma genitalium. The assignment MATERIALS AND METHODS was carried out by our computer server (http:yywww.doe- mbi.ucla.eduypeopleyfrsvryfrsvr.html), which assigns folds to The MG Sequences. The 468 MG sequences were obtained amino acid sequences by comparing sequence-derived predic- from The Institute for Genome Research (TIGR) through its tions with known structures. Of the total of 468 protein ORFs, Web address: http:yywww.tigr.orgytdbymdbymgdbymgd- 103 (22%) can be assigned a known protein fold with high b.html. Three types of annotation (based on searches in the confidence, as cross-validated with tests on known structures. sequence database) accompany each TIGR sequence (10): (i) Of these sequences, 75 (16%) show enough sequence similarity functional assignment—a clear sequence similarity with a to proteins of known structure that they can also be detected protein of known function from another organism was found by traditional sequence–sequence comparison methods. -

Practical Structure-Sequence Alignment of Pseudoknotted Rnas Wei Wang
Practical structure-sequence alignment of pseudoknotted RNAs Wei Wang To cite this version: Wei Wang. Practical structure-sequence alignment of pseudoknotted RNAs. Bioinformatics [q- bio.QM]. Université Paris Saclay (COmUE), 2017. English. NNT : 2017SACLS563. tel-01697889 HAL Id: tel-01697889 https://tel.archives-ouvertes.fr/tel-01697889 Submitted on 31 Jan 2018 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. 1 NNT : 2017SACLS563 Thèse de doctorat de l’Université Paris-Saclay préparée à L’Université Paris-Sud Ecole doctorale n◦580 (STIC) Sciences et Technologies de l’Information et de la Communication Spécialité de doctorat : Informatique par M. Wei WANG Alignement pratique de structure-séquence d’ARN avec pseudonœuds Thèse présentée et soutenue à Orsay, le 18 Décembre 2017. Composition du Jury : Mme Hélène TOUZET Directrice de Recherche (Présidente) CNRS, Université Lille 1 M. Guillaume FERTIN Professeur (Rapporteur) Université de Nantes M. Jan GORODKIN Professeur (Rapporteur) University of Copenhagen Mme Johanne COHEN Directrice de Recherche (Examinatrice) -

UC San Diego UC San Diego Electronic Theses and Dissertations
UC San Diego UC San Diego Electronic Theses and Dissertations Title Analysis and applications of conserved sequence patterns in proteins Permalink https://escholarship.org/uc/item/5kh9z3fr Author Ie, Tze Way Eugene Publication Date 2007 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA, SAN DIEGO Analysis and Applications of Conserved Sequence Patterns in Proteins A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Computer Science by Tze Way Eugene Ie Committee in charge: Professor Yoav Freund, Chair Professor Sanjoy Dasgupta Professor Charles Elkan Professor Terry Gaasterland Professor Philip Papadopoulos Professor Pavel Pevzner 2007 Copyright Tze Way Eugene Ie, 2007 All rights reserved. The dissertation of Tze Way Eugene Ie is ap- proved, and it is acceptable in quality and form for publication on microfilm: Chair University of California, San Diego 2007 iii DEDICATION This dissertation is dedicated in memory of my beloved father, Ie It Sim (1951–1997). iv TABLE OF CONTENTS Signature Page . iii Dedication . iv Table of Contents . v List of Figures . viii List of Tables . x Acknowledgements . xi Vita, Publications, and Fields of Study . xiii Abstract of the Dissertation . xv 1 Introduction . 1 1.1 Protein Homology Search . 1 1.2 Sequence Comparison Methods . 2 1.3 Statistical Analysis of Protein Motifs . 4 1.4 Motif Finding using Random Projections . 5 1.5 Microbial Gene Finding without Assembly . 7 2 Multi-Class Protein Classification using Adaptive Codes . 9 2.1 Profile-based Protein Classifiers . 14 2.2 Embedding Base Classifiers in Code Space . -

ABSTRACT DATA DRIVEN APPROACHES to IDENTIFY DETERMINANTS of HEART DISEASES and CANCER RESISTANCE Avinash Das Sahu, Doctor Of
ABSTRACT Title of dissertation: DATA DRIVEN APPROACHES TO IDENTIFY DETERMINANTS OF HEART DISEASES AND CANCER RESISTANCE Avinash Das Sahu, Doctor of Philosophy, 2016 Dissertation directed by: Professor Sridhar Hannenhalli Department of Computer Science Cancer and cardio-vascular diseases are the leading causes of death world-wide. Caused by systemic genetic and molecular disruptions in cells, these disorders are the manifestation of profound disturbance of normal cellular homeostasis. People suffering or at high risk for these disorders need early diagnosis and personalized therapeutic intervention. Successful implementation of such clinical measures can significantly improve global health. However, development of effective therapies is hindered by the challenges in identifying genetic and molecular determinants of the onset of diseases; and in cases where therapies already exist, the main challenge is to identify molecular determinants that drive resistance to the therapies. Due to the progress in sequencing technologies, the access to a large genome-wide biolog- ical data is now extended far beyond few experimental labs to the global research community. The unprecedented availability of the data has revolutionized the ca- pabilities of computational researchers, enabling them to collaboratively address the long standing problems from many different perspectives. Likewise, this thesis tackles the two main public health related challenges using data driven approaches. Numerous association studies have been proposed to identify genomic variants that determine disease. However, their clinical utility remains limited due to their inability to distinguish causal variants from associated variants. In the presented thesis, we first propose a simple scheme that improves association studies in su- pervised fashion and has shown its applicability in identifying genomic regulatory variants associated with hypertension. -

Etwork Inference from Vector Representations of Words
RZ 3918 (# ZUR1801-015) 01/09/2018 Computer Science/Mathematics 19 pages Research Report INtERAcT: Interaction Network Inference from Vector Representations of Words Matteo Manica, Roland Mathis, Maria Rodriguez Martinez IBM Research – Zurich 8803 Rüschlikon Switzerland LIMITED DISTRIBUTION NOTICE This report has been submitted for publication outside of IBM and will probably be copyrighted if accepted for publication. It has been issued as a Research Report for early dissemination of its contents. In view of the transfer of copyright to the outside pub- lisher, its distribution outside of IBM prior to publication should be limited to peer communications and specific requests. After outside publication, requests should be filled only by reprints or legally obtained copies (e.g., payment of royalties). Some re- ports are available at http://domino.watson.ibm.com/library/Cyberdig.nsf/home. Research Africa • Almaden • Austin • Australia • Brazil • China • Haifa • India • Ireland • Tokyo • Watson • Zurich INtERAcT: Interaction Network Inference from Vector Representations of Words Matteo Manica1,2,*, Roland Mathis1,*, Mar´ıa Rodr´ıguez Mart´ınez1, y ftte,lth,[email protected] 1 IBM Research Z¨urich, S¨aumerstrasse4, 8803 R¨uschlikon, Switzerland 2 ETH Z¨urich, Institute f¨urMolekulare Systembiologie, Auguste-Piccard-Hof 1 8093, Z¨urich, Switzerland * Shared first authorship y Corresponding author Abstract Background In recent years, the number of biomedical publications made freely available through literature archives is steadfastly growing, resulting in a rich source of untapped new knowledge. Most biomedical facts are however buried in the form of unstructured text, and their exploitation requires expert-knowledge and time-consuming manual curation of published articles. -

Predicting Protein Folding Pathways Using Ensemble Modeling and Sequence Information
Predicting Protein Folding Pathways Using Ensemble Modeling and Sequence Information by David Becerra McGill University School of Computer Science Montr´eal, Qu´ebec, Canada A thesis submitted to McGill University in partial fulfillment of the requirements of the degree of Doctor of Philosophy c David Becerra 2017 Dedicated to my GRANDMOM. Thanks for everything. You are such a strong woman i Acknowledgements My doctoral program has been an amazing adventure with ups and downs. The most valuable fact is that I am a different person with respect to the one who started the program five years ago. I am not better or worst, I am just different. I would like to express my gratitude to all the people who helped me during my time at McGill. This thesis contains only my name as author, but it should have the name of all the people who played an important role in the completion of this thesis (and they are a lot of people). I am certain I would have not reached the end of my thesis without the help, advises and support of all people around me. First at all, I want to thank my country Colombia. Being abroad, I learnt to value my culture, my roots and my south-american positiveness, happiness and resourcefulness. I am really proud of being Colombian and of showing to the world a bit of what our wonderful land has given to us. I would also like to thank all Colombians, because with your taxes (via Colciencia’s funding) I was able to complete this dream. -

Protein Engineering Design & Selection
ISSN 1741-0126 (PRINT) ISSN 1741-0134 (ONLINE) peds protein engineering design & selection protein pedsprotein engineering design & selection Special Issue: Computational Protein Structure Assessment and Design: Part III Methods VOLUME GADIS: Algorithm for designing sequences to achieve target secondary structure profiles of intrinsically disordered proteins protein engineering design & selection 29 | peds Tyler S. Harmon, Michael D. Crabtree, Sarah L. Shammas, Ammon E. Posey, Jane Clarke, and Rohit V. Pappu 339 Original Articles NUMBER Conformational dynamics of cancer-associated MyD88-TIR domain mutant L252P (L265P) allosterically tilts the landscape VOLUME 29 NUMBER 9 2016 | | SEPTEMBER toward homo-dimerization 9 | S Chendi Zhan, Ruxi Qi, Guanghong Wei, Emine Guven-Maiorov, Ruth Nussinov, and Buyong Ma 347 E Efficient laboratory evolution of computationally designed enzymes with low starting activities using fluorescence-activated PT EMBER droplet sorting Special Issue: Computational Protein Structure Assessment and Design: Part III Richard Obexer, Moritz Pott, Cathleen Zeymer, Andrew D. Griffiths, and Donald Hilvert 355 2016 Steric interactions determine side-chain conformations in protein cores D. Caballero, A. Virrueta, C.S. O’Hern, and L. Regan 367 www.peds.oxfordjournals.org An in silico algorithm for identifying stabilizing pockets in proteins: test case, the Y220C mutant of the p53 tumor suppressor protein Dennis Bromley, Matthias R. Bauer, Alan R. Fersht, and Valerie Daggett 377 Structural difficulty index: a reliable measure -

BGI Founder and President Visits EMBO the Stars of Biomedicine The
AUTUMN 2012 ISSUE 22 encounters The 4th EMBO Meeting PAGES 3 AND 10 –12 The stars of biomedicine PAGE 8 ©Karlsruhe Institute of Technology (KIT) ©Karlsruhe (KIT) Institute of Technology Eric | Côte d’Azur | Vence Zaragoza PAGE 4 Focus on partnerships BGI founder and President visits EMBO INTERVIEW Gunnar von Heijne, Director of FEATURE The Milan-based Institute of INTERVIEW Sir Paul Nurse discusses the Center for Biomembrane Research at Molecular Oncology Foundation is expanding science, society, and his new venture, The Stockholm University, explains why life would into Asia to recruit top research talent to Italy. Francis Crick Institute, in an interview at be impossible without biological membranes. The 4th EMBO Meeting in Nice, France. PAGE 9 PAGE 6 PAGE 12 www.embo.org COMMENTARY Inside scientific publishing Scooping protection and rapid publication arlier in the year, we started a series of arti- These and other key procedures have been various steps to ensure rapid handling of the cles in EMBOencounters to inform readers included in a formal description of The EMBO manuscript and to avoid protracted revisions. Eabout some of the developments in scientif- Transparent Publishing Process for all four EMBO EMBO editors rapidly respond to submissions. ic publishing. The previous article described the publications (see Figure 1 and journal websites). The initial editorial decision ensures that only ‘Transparent Review’ process used by The EMBO Here, I describe the processes developed and those manuscripts enter full peer review that Journal, EMBO reports, Molecular Systems Biology used by the EMBO publications that allow have a high chance of acceptance in a timely and EMBO Molecular Medicine. -

Drosophila Melanogaster
ORTHOLOGOUS PAIR TRANSFER AND HYBRID BAYES METHODS TO PREDICT THE PROTEIN-PROTEIN INTERACTION NETWORK OF THE ANOPHELES GAMBIAE MOSQUITOES By Qiuxiang Li SUBMITTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY AT IMPERIAL COLLEGE LONDON SOUTH KENSINGTON, LONDON SW7 2AZ 2008 °c Copyright by Qiuxiang Li, 2008 IMPERIAL COLLEGE LONDON DIVISION OF CELL & MOLECULAR BIOLOGY The undersigned hereby certify that they have read and recommend to the Faculty of Natural Sciences for acceptance a thesis entitled \Orthologous pair transfer and hybrid Bayes methods to predict the protein-protein interaction network of the Anopheles gambiae mosquitoes" by Qiuxiang Li in partial ful¯llment of the requirements for the degree of Doctor of Philosophy. Dated: 2008 Research Supervisors: Professor Andrea Crisanti Professor Stephen H. Muggleton Examining Committee: Dr. Michael Tristem Dr. Tania Dottorini ii IMPERIAL COLLEGE LONDON Date: 2008 Author: Qiuxiang Li Title: Orthologous pair transfer and hybrid Bayes methods to predict the protein-protein interaction network of the Anopheles gambiae mosquitoes Division: Cell & Molecular Biology Degree: Ph.D. Convocation: March Year: 2008 Permission is herewith granted to Imperial College London to circulate and to have copied for non-commercial purposes, at its discretion, the above title upon the request of individuals or institutions. Signature of Author THE AUTHOR RESERVES OTHER PUBLICATION RIGHTS, AND NEITHER THE THESIS NOR EXTENSIVE EXTRACTS FROM IT MAY BE PRINTED OR OTHERWISE REPRODUCED WITHOUT THE AUTHOR'S WRITTEN PERMISSION. THE AUTHOR ATTESTS THAT PERMISSION HAS BEEN OBTAINED FOR THE USE OF ANY COPYRIGHTED MATERIAL APPEARING IN THIS THESIS (OTHER THAN BRIEF EXCERPTS REQUIRING ONLY PROPER ACKNOWLEDGEMENT IN SCHOLARLY WRITING) AND THAT ALL SUCH USE IS CLEARLY ACKNOWLEDGED. -
Bibliography
Bibliography [1] Noam Agmon. Tetrahedral displacement: The molecular mechanism behind the Debye relax- ation in water. Journal of Physical Chemistry, 100(1):1072–1080, 1996. [2] M. P. Allen and D. J. Tildesley. Computer Simulation of Liquids. Oxford Science Publications, Clarendon Press, Oxford, 1989. [3] Patrick Aloy and Robert B. Russell. Interrogating protein interaction networks through struc- tural biology. Proceedings of the National Academy of Sciences, USA, 99:5896–5901, 2002. [4] Claus A.F. Andersen, Arthur G. Palmer, Sren Brunak, and Burkhard Rost. Continuum secondary structure captures protein flexibility. Structure, 10:175–184, 2002. [5] D. E. ANDERSON, J. H. HURLEY, H. NICHOLSON, W. A. BAASE, and B. W. MATTHEWS. Hydrophobic core repacking and aromatic-aromatic interaction in the ther- mostable mutant of T4 lysozyme Ser 117 Phe. Protein Science, 2(8):1285–1290, 1993. → [6] G. Apic, J. Gough, and S. A. Teichmann. An insight into domain combinations. Bioinfor- matics, 17 (Suppl.1):S83–89, 2001. [7] Isaiah T. Arkin, Huafeng Xu, Morten O. Jensen, Eyal Arbely, Estelle R. Bennett, Kevin J. Bowers, Edmond Chow, Ron O. Dror, Michael P. Eastwood, Ravenna Flitman-Tene, Brent A. Gregersen, John L. Klepeis, Istvan Kolossvary, Yibing Shan, and David E. Shaw. Mechanism of Na+/H+ antiporting. Science, 317(5839):799–803, 2007. [8] R. Aurora and G. D. Rose. Helix capping. Protein Science, 7(1):21–38, 1998. [9] Kellar Autumn, Metin Sitti, Yiching A. Liang, Anne M. Peattie, Wendy R. Hansen, Simon Sponberg, Thomas W. Kenny, Ronald Fearing, Jacob N. Israelachvili, and Robert J. Full. Evidence for van der Waals adhesion in gecko setae. -

Digital Biology
Digital Biology L. Ridgway Scott University of Chicago Ariel Fern´andez Rice University Release 0.4 DO NOT DISTRIBUTE October 3, 2014 Draft: October 3, 2014, do not distribute Contents 1 How is biology digital? 1 1.1 Exercises....................................... ... 4 2 Digital rules for proteins 5 2.1 Digitalnatureofmolecules. ........ 5 2.1.1 Digitalnatureofatoms. .... 5 2.1.2 Ionicrules.................................... 9 2.1.3 Carbon/hydrogenrules. .... 9 2.2 Digitalnatureofproteins. ........ 10 2.2.1 Hydrophobicityandhydrophilicity . ....... 11 2.2.2 Solvation =salvation............................... 11 6 2.2.3 Energeticambivalence . .... 12 2.3 Pchemomics...................................... .. 13 2.3.1 Anewtool?.................................... 14 2.3.2 Dataminingdefinition . ... 15 2.3.3 Datamininglens ................................ 15 2.3.4 Hydrogen bonds are orientation-dependent . ......... 16 2.3.5 Whatisananswer?............................... 16 2.4 Multiscalemodels................................ ..... 17 2.5 Hydrophobicinteractions. ....... 19 2.5.1 Solventmediationofelectricforces . ......... 20 2.5.2 Dehydrons..................................... 22 2.5.3 Dynamicsofdehydrons. ... 22 2.5.4 Simulateddynamics ............................. .. 25 2.5.5 Stickinessofdehydrons. ..... 26 2.6 Dehydronswitch .................................. ... 26 2.7 Exercises....................................... ... 27 3 Electrostatic forces 29 3.1 Directbonds..................................... ... 29 3.1.1 Covalentbonds ................................ -
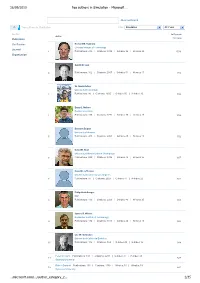
Top Authors in Simulation - Microsoft …
26/09/2010 Top authors in Simulation - Microsoft … Advanced Search Top authors in Simulation Filter: Simulation All Years Author In Domain Author Publication Citations Conference Richard M. Fujimoto Georgia Institute of Technology Journal 1 Publications: 210 | Citations: 3939 | G-Index: 56 | H-Index: 29 1578 Organization Averill M. Law 2 Publications: 102 | Citations: 2657 | G-Index: 51 | H-Index: 17 942 W. David Kelton University of Cincinnati 3 Publications: 86 | Citations: 2605 | G-Index: 50 | H-Index: 15 896 Barry L. Nelson Purdue University 4 Publications: 158 | Citations: 1772 | G-Index: 38 | H-Index: 20 852 Bernard Zeigler University of Arizona 5 Publications: 200 | Citations: 2401 | G-Index: 45 | H-Index: 18 792 David M. Nicol University of Illinois Urbana Champaign 6 Publications: 229 | Citations: 2706 | G-Index: 43 | H-Index: 25 697 David R. Jefferson University of California Los Angeles 7 Publications: 54 | Citations: 2024 | G-Index: 44 | H-Index: 22 681 Philip Heidelberger IBM 8 Publications: 116 | Citations: 2201 | G-Index: 43 | H-Index: 25 603 James R. Wilson Rochester Institute of Technology 9 Publications: 194 | Citations: 1113 | G-Index: 24 | H-Index: 18 586 Lee W. Schruben University of California Berkeley 10 Publications: 102 | Citations: 862 | G-Index: 25 | H-Index: 16 544 Peter W. Glynn Publications: 183 | Citations: 2291 | G-Index: 41 | H-Index: 23 11 527 Stanford University Robert Sargent Publications: 101 | Citations: 1136 | G-Index: 30 | H-Index: 19 12 521 Syracuse University …microsoft.com/…/author_category_2… 1/35 26/09/2010 Top authors in Simulation - Microsoft … Osman Balci Publications: 78 | Citations: 782 | G-Index: 23 | H-Index: 16 13 454 Virginia Polytechnic Institute and State University Jayadev Misra Publications: 136 | Citations: 3325 | G-Index: 56 | H-Index: 26 14 453 University of Texas Austin Rajive Bagrodia Publications: 175 | Citations: 3160 | G-Index: 51 | H-Index: 24 15 435 University of California Los Angeles Richard E.