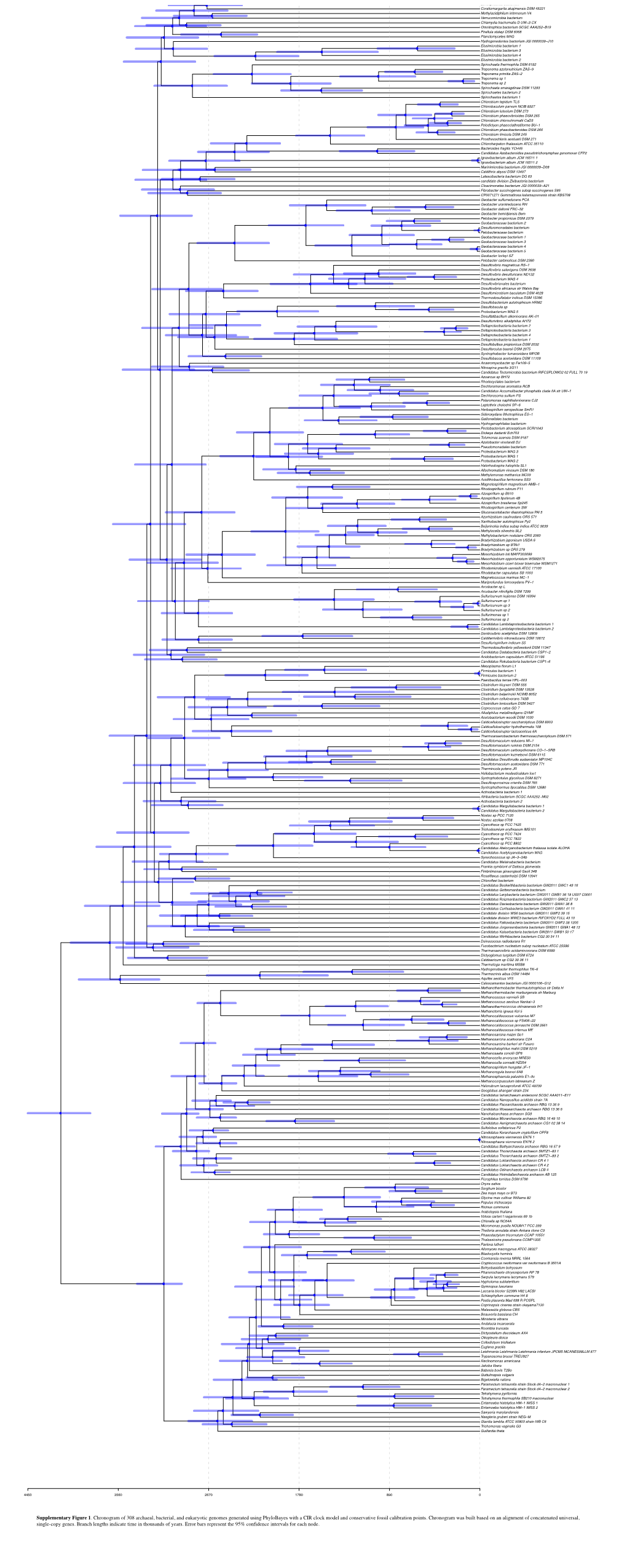
Supplementary Figure 1. Chronogram of 308 Archaeal, Bacterial
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The 2014 Golden Gate National Parks Bioblitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event
National Park Service U.S. Department of the Interior Natural Resource Stewardship and Science The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 ON THIS PAGE Photograph of BioBlitz participants conducting data entry into iNaturalist. Photograph courtesy of the National Park Service. ON THE COVER Photograph of BioBlitz participants collecting aquatic species data in the Presidio of San Francisco. Photograph courtesy of National Park Service. The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 Elizabeth Edson1, Michelle O’Herron1, Alison Forrestel2, Daniel George3 1Golden Gate Parks Conservancy Building 201 Fort Mason San Francisco, CA 94129 2National Park Service. Golden Gate National Recreation Area Fort Cronkhite, Bldg. 1061 Sausalito, CA 94965 3National Park Service. San Francisco Bay Area Network Inventory & Monitoring Program Manager Fort Cronkhite, Bldg. 1063 Sausalito, CA 94965 March 2016 U.S. Department of the Interior National Park Service Natural Resource Stewardship and Science Fort Collins, Colorado The National Park Service, Natural Resource Stewardship and Science office in Fort Collins, Colorado, publishes a range of reports that address natural resource topics. These reports are of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Report Series is used to disseminate comprehensive information and analysis about natural resources and related topics concerning lands managed by the National Park Service. -

Download This Publication (PDF File)
PUBLIC LIBRARY of SCIENCE | plosgenetics.org | ISSN 1553-7390 | Volume 2 | Issue 12 | DECEMBER 2006 GENETICS PUBLIC LIBRARY of SCIENCE www.plosgenetics.org Volume 2 | Issue 12 | DECEMBER 2006 Interview Review Knight in Common Armor: 1949 Unraveling the Genetics 1956 An Interview with Sir John Sulston e225 of Human Obesity e188 Jane Gitschier David M. Mutch, Karine Clément Research Articles Natural Variants of AtHKT1 1964 The Complete Genome 2039 Enhance Na+ Accumulation e210 Sequence and Comparative e206 in Two Wild Populations of Genome Analysis of the High Arabidopsis Pathogenicity Yersinia Ana Rus, Ivan Baxter, enterocolitica Strain 8081 Balasubramaniam Muthukumar, Nicholas R. Thomson, Sarah Jeff Gustin, Brett Lahner, Elena Howard, Brendan W. Wren, Yakubova, David E. Salt Matthew T. G. Holden, Lisa Crossman, Gregory L. Challis, About the Cover Drosophila SPF45: A Bifunctional 1974 Carol Churcher, Karen The jigsaw image of representatives Protein with Roles in Both e178 Mungall, Karen Brooks, Tracey of various lines of eukaryote evolution Splicing and DNA Repair Chillingworth, Theresa Feltwell, refl ects the current lack of consensus as Ahmad Sami Chaouki, Helen K. Zahra Abdellah, Heidi Hauser, to how the major branches of eukaryotes Salz Kay Jagels, Mark Maddison, fi t together. The illustrations from upper Sharon Moule, Mandy Sanders, left to bottom right are as follows: a single Mammalian Small Nucleolar 1984 Sally Whitehead, Michael A. scale from the surface of Umbellosphaera; RNAs Are Mobile Genetic e205 Quail, Gordon Dougan, Julian Amoeba, the large amoeboid organism Elements Parkhill, Michael B. Prentice used as an introduction to protists for Michel J. Weber many school children; Euglena, the iconic Low Levels of Genetic 2052 fl agellate that is often used to challenge Soft Sweeps III: The Signature 1998 Divergence across e215 ideas of plants (Euglena has chloroplasts) of Positive Selection from e186 Geographically and and animals (Euglena moves); Stentor, Recurrent Mutation Linguistically Diverse one of the larger ciliates; Cacatua, the Pleuni S. -

Broadly Sampled Multigene Analyses Yield a Well-Resolved Eukaryotic Tree of Life
Smith ScholarWorks Biological Sciences: Faculty Publications Biological Sciences 10-1-2010 Broadly Sampled Multigene Analyses Yield a Well-Resolved Eukaryotic Tree of Life Laura Wegener Parfrey University of Massachusetts Amherst Jessica Grant Smith College Yonas I. Tekle Smith College Erica Lasek-Nesselquist Marine Biological Laboratory Hilary G. Morrison Marine Biological Laboratory See next page for additional authors Follow this and additional works at: https://scholarworks.smith.edu/bio_facpubs Part of the Biology Commons Recommended Citation Parfrey, Laura Wegener; Grant, Jessica; Tekle, Yonas I.; Lasek-Nesselquist, Erica; Morrison, Hilary G.; Sogin, Mitchell L.; Patterson, David J.; and Katz, Laura A., "Broadly Sampled Multigene Analyses Yield a Well-Resolved Eukaryotic Tree of Life" (2010). Biological Sciences: Faculty Publications, Smith College, Northampton, MA. https://scholarworks.smith.edu/bio_facpubs/126 This Article has been accepted for inclusion in Biological Sciences: Faculty Publications by an authorized administrator of Smith ScholarWorks. For more information, please contact [email protected] Authors Laura Wegener Parfrey, Jessica Grant, Yonas I. Tekle, Erica Lasek-Nesselquist, Hilary G. Morrison, Mitchell L. Sogin, David J. Patterson, and Laura A. Katz This article is available at Smith ScholarWorks: https://scholarworks.smith.edu/bio_facpubs/126 Syst. Biol. 59(5):518–533, 2010 c The Author(s) 2010. Published by Oxford University Press, on behalf of the Society of Systematic Biologists. All rights reserved. For Permissions, please email: [email protected] DOI:10.1093/sysbio/syq037 Advance Access publication on July 23, 2010 Broadly Sampled Multigene Analyses Yield a Well-Resolved Eukaryotic Tree of Life LAURA WEGENER PARFREY1,JESSICA GRANT2,YONAS I. TEKLE2,6,ERICA LASEK-NESSELQUIST3,4, 3 3 5 1,2, HILARY G. -
Species Tree Inference and Update on Very Large Datasets Using Approximation, Randomization, Parallelization, and Vectorization
Species tree inference and update on very large datasets using approximation, randomization, parallelization, and vectorization Siavash Mirarab Electrical and Computer Engineering University of California at San Diego 1 Phylogenetic reconstruction from data Gorilla Human Chimpanzee Orangutan ACTGCACACCG ACTGCCCCCG AATGCCCCCG CTGCACACGG 2 Phylogenetic reconstruction from data CTGCACACCG CTGCACACCG CTGCACACGG Gorilla Human Chimpanzee Orangutan ACTGCACACCG ACTGCCCCCG AATGCCCCCG CTGCACACGG 2 Phylogenetic reconstruction from data CTGCACACCG CTGCACACCG CTGCACACGG Gorilla Human Chimpanzee Orangutan ACTGCACACCG ACTGCCCCCG AATGCCCCCG CTGCACACGG 2 Phylogenetic reconstruction from data CTGCACACCG CTGCACACCG CTGCACACGG Gorilla Human Chimpanzee Orangutan ACTGCACACCG ACTGCCCCCG AATGCCCCCG CTGCACACGG Gorilla ACTGCACACCG Human ACTGC-CCCCG Chimpanzee AATGC-CCCCG Orangutan -CTGCACACGG D 2 Phylogenetic reconstruction from data CTGCACACCG CTGCACACCG CTGCACACGG Gorilla Human Chimpanzee Orangutan ACTGCACACCG ACTGCCCCCG AATGCCCCCG CTGCACACGG Orangutan Chimpanzee Gorilla ACTGCACACCG Human ACTGC-CCCCG Chimpanzee AATGC-CCCCG Orangutan -CTGCACACGG Gorilla Human D P (D T ) T | 2 Applications: HIV forensic Texas case Washington case Scaduto et al., PNAS, 2010 3 Applications: microbiome https://www.nytimes.com/2017/11/06/well/live/ unlocking-the-secrets-of-the-microbiome.html 4 Applications: microbiome https://www.nytimes.com/2017/11/06/well/live/ unlocking-the-secrets-of-the-microbiome.html Morgan, Xochitl C., Nicola Segata, and Curtis Huttenhower. "Trends in genetics (2013) 4 Applications: food safety Tracking the source of a listeriosis outbreak Jackson, Brendan R., et al. Reviews of Infectious Diseases (2016) 5 Fig. 3. Molecular dating of the 2014 outbreak. (A) BEAST dating of the separation of the 2014 lineage from Middle African lineages (SL = Sierra Leone; GN = Guinea; DRC = Democratic Republic of Congo; tMRCA: Sep 2004, 95% HPD: Oct 2002 - May 2006). -

Multigene Eukaryote Phylogeny Reveals the Likely Protozoan Ancestors of Opis- Thokonts (Animals, Fungi, Choanozoans) and Amoebozoa
Accepted Manuscript Multigene eukaryote phylogeny reveals the likely protozoan ancestors of opis- thokonts (animals, fungi, choanozoans) and Amoebozoa Thomas Cavalier-Smith, Ema E. Chao, Elizabeth A. Snell, Cédric Berney, Anna Maria Fiore-Donno, Rhodri Lewis PII: S1055-7903(14)00279-6 DOI: http://dx.doi.org/10.1016/j.ympev.2014.08.012 Reference: YMPEV 4996 To appear in: Molecular Phylogenetics and Evolution Received Date: 24 January 2014 Revised Date: 2 August 2014 Accepted Date: 11 August 2014 Please cite this article as: Cavalier-Smith, T., Chao, E.E., Snell, E.A., Berney, C., Fiore-Donno, A.M., Lewis, R., Multigene eukaryote phylogeny reveals the likely protozoan ancestors of opisthokonts (animals, fungi, choanozoans) and Amoebozoa, Molecular Phylogenetics and Evolution (2014), doi: http://dx.doi.org/10.1016/ j.ympev.2014.08.012 This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain. 1 1 Multigene eukaryote phylogeny reveals the likely protozoan ancestors of opisthokonts 2 (animals, fungi, choanozoans) and Amoebozoa 3 4 Thomas Cavalier-Smith1, Ema E. Chao1, Elizabeth A. Snell1, Cédric Berney1,2, Anna Maria 5 Fiore-Donno1,3, and Rhodri Lewis1 6 7 1Department of Zoology, University of Oxford, South Parks Road, Oxford OX1 3PS, UK. -

Supplementary Information for Microbial Electrochemical Systems Outperform Fixed-Bed Biofilters for Cleaning-Up Urban Wastewater
Electronic Supplementary Material (ESI) for Environmental Science: Water Research & Technology. This journal is © The Royal Society of Chemistry 2016 Supplementary information for Microbial Electrochemical Systems outperform fixed-bed biofilters for cleaning-up urban wastewater AUTHORS: Arantxa Aguirre-Sierraa, Tristano Bacchetti De Gregorisb, Antonio Berná, Juan José Salasc, Carlos Aragónc, Abraham Esteve-Núñezab* Fig.1S Total nitrogen (A), ammonia (B) and nitrate (C) influent and effluent average values of the coke and the gravel biofilters. Error bars represent 95% confidence interval. Fig. 2S Influent and effluent COD (A) and BOD5 (B) average values of the hybrid biofilter and the hybrid polarized biofilter. Error bars represent 95% confidence interval. Fig. 3S Redox potential measured in the coke and the gravel biofilters Fig. 4S Rarefaction curves calculated for each sample based on the OTU computations. Fig. 5S Correspondence analysis biplot of classes’ distribution from pyrosequencing analysis. Fig. 6S. Relative abundance of classes of the category ‘other’ at class level. Table 1S Influent pre-treated wastewater and effluents characteristics. Averages ± SD HRT (d) 4.0 3.4 1.7 0.8 0.5 Influent COD (mg L-1) 246 ± 114 330 ± 107 457 ± 92 318 ± 143 393 ± 101 -1 BOD5 (mg L ) 136 ± 86 235 ± 36 268 ± 81 176 ± 127 213 ± 112 TN (mg L-1) 45.0 ± 17.4 60.6 ± 7.5 57.7 ± 3.9 43.7 ± 16.5 54.8 ± 10.1 -1 NH4-N (mg L ) 32.7 ± 18.7 51.6 ± 6.5 49.0 ± 2.3 36.6 ± 15.9 47.0 ± 8.8 -1 NO3-N (mg L ) 2.3 ± 3.6 1.0 ± 1.6 0.8 ± 0.6 1.5 ± 2.0 0.9 ± 0.6 TP (mg -

Molecular Identification and Evolution of Protozoa Belonging to the Parabasalia Group and the Genus Blastocystis
UNIVERSITAR DEGLI STUDI DI SASSARI SCUOLA DI DOTTORATO IN SCIENZE BIOMOLECOLARI E BIOTECNOLOGICHE (Intenational PhD School in Biomolecular and Biotechnological Sciences) Indirizzo: Microbiologia molecolare e clinica Molecular identification and evolution of protozoa belonging to the Parabasalia group and the genus Blastocystis Direttore della scuola: Prof. Masala Bruno Relatore: Prof. Pier Luigi Fiori Correlatore: Dott. Eric Viscogliosi Tesi di Dottorato : Dionigia Meloni XXIV CICLO Nome e cognome: Dionigia Meloni Titolo della tesi : Molecular identification and evolution of protozoa belonging to the Parabasalia group and the genus Blastocystis Tesi di dottorato in scienze Biomolecolari e biotecnologiche. Indirizzo: Microbiologia molecolare e clinica Universit degli studi di Sassari UNIVERSITAR DEGLI STUDI DI SASSARI SCUOLA DI DOTTORATO IN SCIENZE BIOMOLECOLARI E BIOTECNOLOGICHE (Intenational PhD School in Biomolecular and Biotechnological Sciences) Indirizzo: Microbiologia molecolare e clinica Molecular identification and evolution of protozoa belonging to the Parabasalia group and the genus Blastocystis Direttore della scuola: Prof. Masala Bruno Relatore: Prof. Pier Luigi Fiori Correlatore: Dott. Eric Viscogliosi Tesi di Dottorato : Dionigia Meloni XXIV CICLO Nome e cognome: Dionigia Meloni Titolo della tesi : Molecular identification and evolution of protozoa belonging to the Parabasalia group and the genus Blastocystis Tesi di dottorato in scienze Biomolecolari e biotecnologiche. Indirizzo: Microbiologia molecolare e clinica Universit degli studi di Sassari Abstract My thesis was conducted on the study of two groups of protozoa: the Parabasalia and Blastocystis . The first part of my work was focused on the identification, pathogenicity, and phylogeny of parabasalids. We showed that Pentatrichomonas hominis is a possible zoonotic species with a significant potential of transmission by the waterborne route and could be the aetiological agent of gastrointestinal troubles in children. -

Metagenomes of Native and Electrode-Enriched Microbial Communities from the Soudan Iron Mine Jonathan P
Metagenomes of native and electrode-enriched microbial communities from the Soudan Iron Mine Jonathan P. Badalamenti and Daniel R. Bond Department of Microbiology and BioTechnology Institute, University of Minnesota - Twin Cities, Saint Paul, Minnesota, USA Twitter: @JonBadalamenti @wanderingbond Summary Approach - compare metagenomes from native and electrode-enriched deep subsurface microbial communities 30 Despite apparent carbon limitation, anoxic deep subsurface brines at the Soudan ) enriched 2 Underground Iron Mine harbor active microbial communities . To characterize these 20 A/cm µ assemblages, we performed shotgun metagenomics of native and enriched samples. enrich ( harvest cells collect inoculate +0.24 V extract 10 Follwing enrichment on poised electrodes and long read sequencing, we recovered Soudan brine electrode 20° C from DNA biodreactors current electrodes from the metagenome the closed, circular genome of a novel Desulfuromonas sp. 0 0 10 20 30 40 filtrate PacBio RS II Illumina HiSeq with remarkable genomic features that were not fully resolved by short read assem- extract time (d) long reads short reads TFF DNA unenriched bly alone. This organism was essentially absent in unenriched Soudan communities, 0.1 µm retentate assembled metagenomes reconstruct long read return de novo complete genome(s) assembly indicating that electrodes are highly selective for putative metal reducers. Native HGAP assembly community metagenomes suggest that carbon cycling is driven by methyl-C me- IDBA_UD 1 hybrid tabolism, in particular methylotrophic methanogenesis. Our results highlight the 3 µm prefilter assembly N4 binning promising potential for long reads in metagenomic surveys of low-diversity environ- borehole N4 binning read trimming and filtering brine de novo ments. -

Author's Manuscript (764.7Kb)
1 BROADLY SAMPLED TREE OF EUKARYOTIC LIFE Broadly Sampled Multigene Analyses Yield a Well-resolved Eukaryotic Tree of Life Laura Wegener Parfrey1†, Jessica Grant2†, Yonas I. Tekle2,6, Erica Lasek-Nesselquist3,4, Hilary G. Morrison3, Mitchell L. Sogin3, David J. Patterson5, Laura A. Katz1,2,* 1Program in Organismic and Evolutionary Biology, University of Massachusetts, 611 North Pleasant Street, Amherst, Massachusetts 01003, USA 2Department of Biological Sciences, Smith College, 44 College Lane, Northampton, Massachusetts 01063, USA 3Bay Paul Center for Comparative Molecular Biology and Evolution, Marine Biological Laboratory, 7 MBL Street, Woods Hole, Massachusetts 02543, USA 4Department of Ecology and Evolutionary Biology, Brown University, 80 Waterman Street, Providence, Rhode Island 02912, USA 5Biodiversity Informatics Group, Marine Biological Laboratory, 7 MBL Street, Woods Hole, Massachusetts 02543, USA 6Current address: Department of Epidemiology and Public Health, Yale University School of Medicine, New Haven, Connecticut 06520, USA †These authors contributed equally *Corresponding author: L.A.K - [email protected] Phone: 413-585-3825, Fax: 413-585-3786 Keywords: Microbial eukaryotes, supergroups, taxon sampling, Rhizaria, systematic error, Excavata 2 An accurate reconstruction of the eukaryotic tree of life is essential to identify the innovations underlying the diversity of microbial and macroscopic (e.g. plants and animals) eukaryotes. Previous work has divided eukaryotic diversity into a small number of high-level ‘supergroups’, many of which receive strong support in phylogenomic analyses. However, the abundance of data in phylogenomic analyses can lead to highly supported but incorrect relationships due to systematic phylogenetic error. Further, the paucity of major eukaryotic lineages (19 or fewer) included in these genomic studies may exaggerate systematic error and reduces power to evaluate hypotheses. -

Uneven Distribution of Cobamide Biosynthesis and Dependence in Bacteria Predicted by Comparative Genomics
The ISME Journal (2018) 13:789–804 https://doi.org/10.1038/s41396-018-0304-9 ARTICLE Uneven distribution of cobamide biosynthesis and dependence in bacteria predicted by comparative genomics 1 1 1 1,2 3,4 3 Amanda N. Shelton ● Erica C. Seth ● Kenny C. Mok ● Andrew W. Han ● Samantha N. Jackson ● David R. Haft ● Michiko E. Taga1 Received: 7 June 2018 / Revised: 14 September 2018 / Accepted: 4 October 2018 / Published online: 14 November 2018 © The Author(s) 2018. This article is published with open access Abstract The vitamin B12 family of cofactors known as cobamides are essential for a variety of microbial metabolisms. We used comparative genomics of 11,000 bacterial species to analyze the extent and distribution of cobamide production and use across bacteria. We find that 86% of bacteria in this data set have at least one of 15 cobamide-dependent enzyme families, but only 37% are predicted to synthesize cobamides de novo. The distribution of cobamide biosynthesis and use vary at the phylum level. While 57% of Actinobacteria are predicted to biosynthesize cobamides, only 0.6% of Bacteroidetes have the complete pathway, yet 96% of species in this phylum have cobamide-dependent enzymes. The form of cobamide produced 1234567890();,: 1234567890();,: by the bacteria could be predicted for 58% of cobamide-producing species, based on the presence of signature lower ligand biosynthesis and attachment genes. Our predictions also revealed that 17% of bacteria have partial biosynthetic pathways, yet have the potential to salvage cobamide precursors. Bacteria with a partial cobamide biosynthesis pathway include those in a newly defined, experimentally verified category of bacteria lacking the first step in the biosynthesis pathway. -

Synchronized Dynamics of Bacterial Nichespecific Functions During
Synchronized dynamics of bacterial niche-specific functions during biofilm development in a cold seep brine pool Item Type Article Authors Zhang, Weipeng; Wang, Yong; Bougouffa, Salim; Tian, Renmao; Cao, Huiluo; Li, Yongxin; Cai, Lin; Wong, Yue Him; Zhang, Gen; Zhou, Guowei; Zhang, Xixiang; Bajic, Vladimir B.; Al-Suwailem, Abdulaziz M.; Qian, Pei-Yuan Citation Synchronized dynamics of bacterial niche-specific functions during biofilm development in a cold seep brine pool 2015:n/a Environmental Microbiology Eprint version Post-print DOI 10.1111/1462-2920.12978 Publisher Wiley Journal Environmental Microbiology Rights This is the peer reviewed version of the following article: Zhang, Weipeng, Yong Wang, Salim Bougouffa, Renmao Tian, Huiluo Cao, Yongxin Li, Lin Cai et al. "Synchronized dynamics of bacterial niche-specific functions during biofilm development in a cold seep brine pool." Environmental microbiology (2015)., which has been published in final form at http:// doi.wiley.com/10.1111/1462-2920.12978. This article may be used for non-commercial purposes in accordance With Wiley Terms and Conditions for self-archiving. Download date 25/09/2021 02:54:27 Link to Item http://hdl.handle.net/10754/561085 Synchronized dynamics of bacterial niche-specific functions during biofilm development in a cold seep brine pool1 Weipeng Zhang1, Yong Wang2, Salim Bougouffa3, Renmao Tian1, Huiluo Cao1, Yongxin Li1 Lin Cai1, Yue Him Wong1, Gen Zhang1, Guowei Zhou1, Xixiang Zhang3, Vladimir B Bajic3, Abdulaziz Al-Suwailem3, Pei-Yuan Qian1,2# 1KAUST Global -

Development of UASB-DHS System for Treating Industrial Wastewater Containing Ethylene Glycol
Journal of Water and Environment Technology, Vol.13, No.2, 2015 Development of UASB-DHS System for Treating Industrial Wastewater Containing Ethylene Glycol Takahiro WATARI 1), Daisuke TANIKAWA2), Kyohei KURODA1), Akinobu NAKAMURA1), Nanako FUJII3), Fuminori YONEYAMA3), Osamu WAKISAKA3), Masashi HATAMOTO1), Takashi YAMAGUCHI 1) 1) Department of Environmental Systems Engineering, Nagaoka University of Technology, 1603-1 Kamitomioka, Nagaoka, Niigata 940-2188, Japan 2) Department of Civil and Environmental Engineering, National Institute of Technology, Kure College, 2-1-11 Agaminami, Kure, Hiroshima 737-8506, Japan 3) Fundamental Material Laboratory Office, Material Technology Research and Development Laboratory, Research and Development Headquarters, Sumitomo Riko Company Limited, 3-1 Higashi, Komaki, Aichi 485-8550, Japan ABSTRACT This study evaluated the performance of a novel treatment system consisting of an upflow anaerobic sludge blanket (UASB) and a downflow hanging sponge (DHS) for the treatment of industrial wastewater containing 8% ethylene glycol and 2% propylene glycol discharged from a rubber production unit. The system achieved high COD removal (91 ± 4.3%) and methane recovery (82 ± 20%) at an organic loading rate of 8.5 kg-COD/(m3·day). The UASB allowed an organic loading rate of 14 kg-COD/(m3·day) with a constant hydraulic retention time of 24 h. The COD of DHS effluent was 370 ± 250 mg-COD/L during the entire experimental period. Thus, the proposed system could be applicable for treating industrial wastewater containing ethylene glycol. Massively parallel 16S rRNA gene sequencing elucidated the microbial community structure of the UASB. The dominant family Pelobacteriaceae could mainly degrade the organic compounds of ethylene glycol and decomposed products of ethanol.