Comparative Chloroplast Genomics of Endangered Euphorbia Species: Insights Into Hotspot Divergence, Repetitive Sequence Variation, and Phylogeny
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Proceedings Amurga Co
PROCEEDINGS OF THE AMURGA INTERNATIONAL CONFERENCES ON ISLAND BIODIVERSITY 2011 PROCEEDINGS OF THE AMURGA INTERNATIONAL CONFERENCES ON ISLAND BIODIVERSITY 2011 Coordination: Juli Caujapé-Castells Funded and edited by: Fundación Canaria Amurga Maspalomas Colaboration: Faro Media Cover design & layout: Estudio Creativo Javier Ojeda © Fundación Canaria Amurga Maspalomas Gran Canaria, December 2013 ISBN: 978-84-616-7394-0 How to cite this volume: Caujapé-Castells J, Nieto Feliner G, Fernández Palacios JM (eds.) (2013) Proceedings of the Amurga international conferences on island biodiversity 2011. Fundación Canaria Amurga-Maspalomas, Las Palmas de Gran Canaria, Spain. All rights reserved. Any unauthorized reprint or use of this material is prohibited. No part of this book may be reproduced or transmitted in any form or by any means, electronic or mechanical, including photocopying, recording, or by any information storage and retrieval system without express written permission from the author / publisher. SCIENTIFIC EDITORS Juli Caujapé-Castells Jardín Botánico Canario “Viera y Clavijo” - Unidad Asociada CSIC Consejería de Medio Ambiente y Emergencias, Cabildo de Gran Canaria Gonzalo Nieto Feliner Real Jardín Botánico de Madrid-CSIC José María Fernández Palacios Universidad de La Laguna SCIENTIFIC COMMITTEE Juli Caujapé-Castells, Gonzalo Nieto Feliner, David Bramwell, Águedo Marrero Rodríguez, Julia Pérez de Paz, Bernardo Navarro-Valdivielso, Ruth Jaén-Molina, Rosa Febles Hernández, Pablo Vargas. Isabel Sanmartín. ORGANIZING COMMITTEE Pedro -

Euphorbia Telephioides (Euphorbiaceae)
Genetic diversity within a threatened, endemic North American species, Euphorbia telephioides (Euphorbiaceae) Dorset W. Trapnell, J. L. Hamrick & Vivian Negrón-Ortiz Conservation Genetics ISSN 1566-0621 Conserv Genet DOI 10.1007/s10592-012-0323-4 1 23 Your article is protected by copyright and all rights are held exclusively by Springer Science+Business Media B.V.. This e-offprint is for personal use only and shall not be self- archived in electronic repositories. If you wish to self-archive your work, please use the accepted author’s version for posting to your own website or your institution’s repository. You may further deposit the accepted author’s version on a funder’s repository at a funder’s request, provided it is not made publicly available until 12 months after publication. 1 23 Author's personal copy Conserv Genet DOI 10.1007/s10592-012-0323-4 RESEARCH ARTICLE Genetic diversity within a threatened, endemic North American species, Euphorbia telephioides (Euphorbiaceae) Dorset W. Trapnell • J. L. Hamrick • Vivian Negro´n-Ortiz Received: 23 September 2011 / Accepted: 20 January 2012 Ó Springer Science+Business Media B.V. 2012 Abstract The southeastern United States and Florida which it occurs, Gulf (0.084), Franklin (0.059) and Bay support an unusually large number of endemic plant spe- Counties (0.033), were also quite low. Peripheral popula- cies, many of which are threatened by anthropogenic tions did not generally have reduced genetic variation habitat disturbance. As conservation measures are under- although there was significant isolation by distance. Rare- taken and recovery plans designed, a factor that must be faction analysis showed a non-significant relationship taken into consideration is the genetic composition of the between allelic richness and actual population sizes. -

Euphorbia Subg
ФЕДЕРАЛЬНОЕ ГОСУДАРСТВЕННОЕ БЮДЖЕТНОЕ УЧРЕЖДЕНИЕ НАУКИ БОТАНИЧЕСКИЙ ИНСТИТУТ ИМ. В.Л. КОМАРОВА РОССИЙСКОЙ АКАДЕМИИ НАУК На правах рукописи Гельтман Дмитрий Викторович ПОДРОД ESULA РОДА EUPHORBIA (EUPHORBIACEAE): СИСТЕМА, ФИЛОГЕНИЯ, ГЕОГРАФИЧЕСКИЙ АНАЛИЗ 03.02.01 — ботаника ДИССЕРТАЦИЯ на соискание ученой степени доктора биологических наук САНКТ-ПЕТЕРБУРГ 2015 2 Оглавление Введение ......................................................................................................................................... 3 Глава 1. Род Euphorbia и основные проблемы его систематики ......................................... 9 1.1. Общая характеристика и систематическое положение .......................................... 9 1.2. Краткая история таксономического изучения и формирования системы рода ... 10 1.3. Основные проблемы систематики рода Euphorbia и его подрода Esula на рубеже XX–XXI вв. и пути их решения ..................................................................................... 15 Глава 2. Материал и методы исследования ........................................................................... 17 Глава 3. Построение системы подрода Esula рода Euphorbia на основе молекулярно- филогенетического подхода ...................................................................................................... 24 3.1. Краткая история молекулярно-филогенетического изучения рода Euphorbia и его подрода Esula ......................................................................................................... 24 3.2. Результаты молекулярно-филогенетического -

Petition for the Release of Aphthona Cyparissiae Against Leafy Spurge in the United States1
Reprinted with permission from: USDA-ARS, March 19, 1986, unpublished. Petition for the release of Aphthona cyparissiae against leafy spurge in the 1 United States ROBERT W. PEMBERTON Part I TO: Dr. R. Bovey, Dept. of Range Science USDA/ARS/SR Texas A&M University College Station, TX 77843 U.S.A. Enclosed are the results of the research on the flea beetle Aphthona cyparissiae (Chrysomelidae). This petition is composed of two parts. The first is the report “Aph- thona cyparissiae (Kock) and A. flava Guill. (Coleopera: Chrysomelidae): Two candi- dates for the biological control of cypress and leafy spurge in North America” by G. Sommer and E. Maw which the Working Group reviewed on Canada’s behalf. Copies of this report should be in the Working Group’s files. This research, which was done at the Commonwealth Institute of Biological Control’s Delémont, Switzerland lab, showed A. cyparissiae to be specific to the genus Euphorbia of the Euphorbiaceae. This result agrees with the literature and field records for A. cyparissiae, which recorded it from: Euphorbia cyparissias, E. esula, E. peplus, E. seguieriana, and E. virgata. The Sommer and Maw report also included information on A. cyparissiae’s taxonomic position, life history, laboratory biology, mortality factors, feeding effects on the host plants, the Harris scoring system, as well as a brief description of the target plant – leafy spurge (Euphorbia esula complex), a serious pest of the rangelands of the Great Plains of North America. Based on this research, the Working Group gave permission to release A. cyparissiae in Canada and to import it into the USDA’s Biological Control of Weeds Quarantine in Al- bany, California, for additional testing. -

Cataleg Biodiversitat Albufera Mallorca 1998
Edita: Conselleria de Medi Ambient Direcció General de Biodiversitat Dibuix de la coberta: Joan Miquel Bennàssar Impressió: Gràfiques Son Espanyolet Diposit Legal: PM 1992-2002 Pròleg Aquesta obra que teniu a les mans és el producte d’un llarg procés de recerca i d’investiga- ció. És el resultat de més de 12 anys de treballs que han duit a terme els científics del grup s‘Albufera-IBG i molts d‘altres experts i aficionats que han aportat l’esforç i els coneixe- ments per completar aquesta visió global de la biodiversitat de s‘Albufera de Mallorca. Un cos de coneixements que s’ha anat formant pas a pas, com és característic de tot procés cien- tífic. La Conselleria de Medi Ambient és ben conscient que la conservació del patrimoni natural no es pot fer sense els estudis que ens han de permetre conèixer quins són els recursos sota la nostra responsabilitat i quin valor tenen. De cada vegada el gran públic coneix més quines són les espècies amenaçades i les accions per conservar-les troben ressò en els mitjans de comunicació i en la sensibilitat de la socie- tat en general. Però cal demanar-se: estam fent tot allò que és necessari per garantir la con- servació del patrimoni natural per a les generacions pròximes? Són suficients els coneixe- ments actuals d’aquest patrimoni? Quines espècies o quines poblacions són les més valuo- ses? Aquest llibre ajudarà a respondre algunes d‘aquestes preguntes. És un compromís entre la visió del científic, que sempre desitja aprofundir en l’estudi, i els gestors, que demanen amb urgència unes eines adequades per planificar les accions de conservació i avaluar-les. -
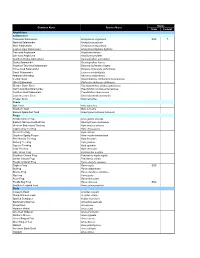
St. Joseph Bay Native Species List
Status Common Name Species Name State Federal Amphibians Salamanders Flatwoods Salamander Ambystoma cingulatum SSC T Marbled Salamander Ambystoma opacum Mole Salamander Ambystoma talpoideum Eastern Tiger Salamander Ambystoma tigrinum tigrinum Two-toed Amphiuma Amphiuma means One-toed Amphiuma Amphiuma pholeter Southern Dusky Salamander Desmognathus auriculatus Dusky Salamander Desmognathus fuscus Southern Two-lined Salamander Eurycea bislineata cirrigera Three-lined Salamander Eurycea longicauda guttolineata Dwarf Salamander Eurycea quadridigitata Alabama Waterdog Necturus alabamensis Central Newt Notophthalmus viridescens louisianensis Slimy Salamander Plethodon glutinosus glutinosus Slender Dwarf Siren Pseudobranchus striatus spheniscus Gulf Coast Mud Salamander Pseudotriton montanus flavissimus Southern Red Salamander Pseudotriton ruber vioscai Eastern Lesser Siren Siren intermedia intermedia Greater Siren Siren lacertina Toads Oak Toad Bufo quercicus Southern Toad Bufo terrestris Eastern Spadefoot Toad Scaphiopus holbrooki holbrooki Frogs Florida Cricket Frog Acris gryllus dorsalis Eastern Narrow-mouthed Frog Gastrophryne carolinensis Western Bird-voiced Treefrog Hyla avivoca avivoca Cope's Gray Treefrog Hyla chrysoscelis Green Treefrog Hyla cinerea Southern Spring Peeper Hyla crucifer bartramiana Pine Woods Treefrog Hyla femoralis Barking Treefrog Hyla gratiosa Squirrel Treefrog Hyla squirella Gray Treefrog Hyla versicolor Little Grass Frog Limnaoedus ocularis Southern Chorus Frog Pseudacris nigrita nigrita Ornate Chorus Frog Pseudacris -

Integrated Noxious Weed Management Plan: US Air Force Academy and Farish Recreation Area, El Paso County, CO
Integrated Noxious Weed Management Plan US Air Force Academy and Farish Recreation Area August 2015 CNHP’s mission is to preserve the natural diversity of life by contributing the essential scientific foundation that leads to lasting conservation of Colorado's biological wealth. Colorado Natural Heritage Program Warner College of Natural Resources Colorado State University 1475 Campus Delivery Fort Collins, CO 80523 (970) 491-7331 Report Prepared for: United States Air Force Academy Department of Natural Resources Recommended Citation: Smith, P., S. S. Panjabi, and J. Handwerk. 2015. Integrated Noxious Weed Management Plan: US Air Force Academy and Farish Recreation Area, El Paso County, CO. Colorado Natural Heritage Program, Colorado State University, Fort Collins, Colorado. Front Cover: Documenting weeds at the US Air Force Academy. Photos courtesy of the Colorado Natural Heritage Program © Integrated Noxious Weed Management Plan US Air Force Academy and Farish Recreation Area El Paso County, CO Pam Smith, Susan Spackman Panjabi, and Jill Handwerk Colorado Natural Heritage Program Warner College of Natural Resources Colorado State University Fort Collins, Colorado 80523 August 2015 EXECUTIVE SUMMARY Various federal, state, and local laws, ordinances, orders, and policies require land managers to control noxious weeds. The purpose of this plan is to provide a guide to manage, in the most efficient and effective manner, the noxious weeds on the US Air Force Academy (Academy) and Farish Recreation Area (Farish) over the next 10 years (through 2025), in accordance with their respective integrated natural resources management plans. This plan pertains to the “natural” portions of the Academy and excludes highly developed areas, such as around buildings, recreation fields, and lawns. -

Extent of Euphorbia Paralias in Identified Neophema Chrysogaster Feeding Grounds in the Arthur-Pieman Conservation Area Between Native Well Bay and Pavement Point
Extent of Euphorbia paralias in identified Neophema chrysogaster feeding grounds in the Arthur-Pieman Conservation Area between Native Well Bay and Pavement Point Priority Sea Spurge Eradication in Identified Arthur-Pieman Conservation Area Orange Bellied Parrot Feeding Grounds Project, funded by the Natural Heritage Trust through the Cradle Coast Regional Weed Management Steering Committee. North-West Environment Centre Inc. 16 April 2007 Extent of Euphorbia paralias in identified Neophema chrysogaster feeding grounds in the Arthur-Pieman Conservation Area between Native Well Bay and Pavement Point North-West Environment Centre Inc. PO Box 999, Burnie Tasmania 7320 Author: Matthew Campbell-Ellis Project title: Priority Sea Spurge Eradication in Identified Arthur-Pieman Conservation Area Orange Bellied Parrot Feeding Grounds Editing: Niko Campbell-Ellis, David Henderson, Peter Sims, and Greg Taylor. Photo credits: Matthew Campbell-Ellis, Peter Sims. Suggested citing: Campbell-Ellis, M. 2007. Extent of Euphorbia paralias in identified Neophema chrysogaster feeding grounds in the Arthur-Pieman Conservation Area between Native Well Bay and Pavement Point. North-West Environment Centre Inc. Burnie, Tasmania. Contents Summary 1 Introduction 1 The Orange-bellied Parrot (Neophema chrysogaster) 1 Sea Spurge (Euphorbia paralias) 2 Methodology 4 Mapping results 5 Priority sites for control 6 Site 1: Pavement Point 7 Site 2: Nettley Bay to West Point 7 Site 3: Arthur River bridge to Australia Point 7 Site 4: Temma to south of Richardson Point 7 Site 5: Southern end of Sandy Cape Beach to the southern end of Native Well Bay 7 Acknowledgements 9 Appendix: Maps 10 References 20 Extent of E. paralias in identified N. -

The Evolutionary Fate of Rpl32 and Rps16 Losses in the Euphorbia Schimperi (Euphorbiaceae) Plastome Aldanah A
www.nature.com/scientificreports OPEN The evolutionary fate of rpl32 and rps16 losses in the Euphorbia schimperi (Euphorbiaceae) plastome Aldanah A. Alqahtani1,2* & Robert K. Jansen1,3 Gene transfers from mitochondria and plastids to the nucleus are an important process in the evolution of the eukaryotic cell. Plastid (pt) gene losses have been documented in multiple angiosperm lineages and are often associated with functional transfers to the nucleus or substitutions by duplicated nuclear genes targeted to both the plastid and mitochondrion. The plastid genome sequence of Euphorbia schimperi was assembled and three major genomic changes were detected, the complete loss of rpl32 and pseudogenization of rps16 and infA. The nuclear transcriptome of E. schimperi was sequenced to investigate the transfer/substitution of the rpl32 and rps16 genes to the nucleus. Transfer of plastid-encoded rpl32 to the nucleus was identifed previously in three families of Malpighiales, Rhizophoraceae, Salicaceae and Passiforaceae. An E. schimperi transcript of pt SOD-1- RPL32 confrmed that the transfer in Euphorbiaceae is similar to other Malpighiales indicating that it occurred early in the divergence of the order. Ribosomal protein S16 (rps16) is encoded in the plastome in most angiosperms but not in Salicaceae and Passiforaceae. Substitution of the E. schimperi pt rps16 was likely due to a duplication of nuclear-encoded mitochondrial-targeted rps16 resulting in copies dually targeted to the mitochondrion and plastid. Sequences of RPS16-1 and RPS16-2 in the three families of Malpighiales (Salicaceae, Passiforaceae and Euphorbiaceae) have high sequence identity suggesting that the substitution event dates to the early divergence within Malpighiales. -

Macbridea Alba
Macbridea alba (White birds-in-a-nest) 5-Year Review: Summary and Evaluation Lathrop Management Area, Bay County. Photos by Vivian Negrón-Ortiz U.S. Fish and Wildlife Service Southeast Region Panama City Field Office Panama City, Florida 5-YEAR REVIEW Macbridea alba (White birds-in-a-nest) I. GENERAL INFORMATION A. Methodology used to complete the review This review was accomplished using information obtained from the plant’s 1994 Recovery Plan, peer reviewed scientific publications, unpublished field survey results, reports of current research projects, unpublished field observations by Service, State and other experienced biologists, and personal communications. These documents are on file at the Panama City Field Office. A Federal Register notice announcing the review and requesting information was published on April 16, 2008 (73 FR 20702). Comments received and suggestions from peer reviewers were evaluated and incorporated as appropriate (see appendix A). No part of this review was contracted to an outside party. This review was completed by the Service’s lead Recovery botanist in the Panama City Field Office, Florida. B. Reviewers Lead Field Office: Dr. Vivian Negrón-Ortiz, Panama City Field Office, 850-769-0552 ext. 231, [email protected] Lead Region: Southeast Region: Kelly Bibb, 404-679-7132 Peer reviewers: Ms. Louise Kirn, District Ecologist Apalachicola National Forest P.O. Box 579, Bristol, FL 32321 Ms. Faye Winters, Field Office Biologist BLM Jackson Field Office 411 Briarwood Drive, Suite 404 Jackson, MS 39206 C. Background 1. FR Notice citation announcing initiation of this review: 73 FR 20702 (April 16, 2008). 1 2. Species status: Unknown (Recovery Data Call 2008); the species status is unknown until all the Element Occurrences1 (EO’s) are revisited. -

Pollen Morphology of the Euphorbiaceae with Special Reference to Taxonomy
Pollen morphology of the Euphorbiaceae with special reference to taxonomy W. Punt (Botanical Museum and Herbarium, Utrecht) {received December 28th, 1961) CONTENTS Chapter I General Introduction 2 a. Introduction 2 b. Acknowledgements 2 Chapter II History 2 a. Pollen morphology 2 b. Euphorbiaceae 4 Chapter III Material 5 Chapter IV Methods 6 a. Flowers 6 b. Pollen preparations 7 c. Preservation of pollen grains 7 d. Microscopes 8 e. Punched cards 8 f. Drawings 9 Chapter V Some nomenclatural remarks 9 Chapter VI Pollen morphology 10 Chapter VII Glossary 15 Chapter VIII Results 18 A. Pollen grains of the Euphorbiaceae 18 B. Discussion of the results 20 a. Phyllanthoideae 20 Antidesma configuration 20 Amanoa configuration 32 Phyllanthus nutans configuration 37 Breynia configuration 38 Aristogeitonia configuration 40 b. Crotonoideae 47 Croton configuration 47 Cnesmosa configuration 57 Dysopsis configuration 60 Plukenetia configuration 60 Chiropetalum configuration 65 Cephalomappa configuration 68 Sumbavia configuration 69 Bernardia configuration 73 Mallotus configuration 77 Claoxylon configuration 90 Cladogynos configuration 93 Hippomane configuration 95 Summary 106 References 107 Index 110 CHAPTER I GENERAL INTRODUCTION a. Introduction Many investigators have stated (e.g. Lindau 1895, Wodehouse Erdtman that 1935, 1952), pollen morphology can be of great also importance for plant taxonomy, while it was known that in Euphorbiaceae several types of pollen grains exist (e.g. Erdtman 1952). On the suggestion of Professor Lanjouw, who himself has worked on the Euphorbiaceae of Surinam, the author has investigated the pollen grains of this family of that area. From the result it was apparent that in the Surinam different could be Euphorbiaceae many pollen types distinguished. -

Chapter 14. Wildlife and Forest Communities 341
chapteR 14. Wildlife and Forest Communities 341 Chapter 14. Wildlife and Forest communities Margaret Trani Griep and Beverly Collins1 key FindingS • Hotspot areas for plants of concern are Big Bend National Park; the Apalachicola area of the Southern Gulf Coast; • The South has 1,076 native terrestrial vertebrates: 179 Lake Wales Ridge and the area south of Lake Okeechobee amphibians, 525 birds, 176 mammals, and 196 reptiles. in Peninsular Florida; and coastal counties of North Species richness is highest in the Mid-South (856) and Carolina in the Atlantic Coastal Plain. The Appalachian- Coastal Plain (733), reflecting both the large area of these Cumberland highlands also contain plants identified by subregions and the diversity of habitats within them. States as species of concern. • The geography of species richness varies by taxa. • Species, including those of conservation concern, are Amphibians flourish in portions of the Piedmont and imperiled by habitat alteration, isolation, introduction of Appalachian-Cumberland highlands and across the Coastal invasive species, environmental pollutants, commercial Plain. Bird richness is highest along the coastal wetlands of development, human disturbance, and exploitation. the Atlantic Ocean and Gulf of Mexico, mammal richness Conditions predicted by the forecasts will magnify these is highest in the Mid-South and Appalachian-Cumberland stressors. Each species varies in its vulnerability to highlands, and reptile richness is highest across the forecasted threats, and these threats vary by subregion. Key southern portion of the region. areas of concern arise where hotspots of vulnerable species • The South has 142 terrestrial vertebrate species coincide with forecasted stressors. considered to be of conservation concern (e.g., global • There are 614 species that are presumed extirpated from conservation status rank of critically imperiled, imperiled, selected States in the South; 64 are terrestrial vertebrates or vulnerable), 77 of which are listed as threatened or and 550 are vascular plants.