Euphorbia Telephioides (Euphorbiaceae)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Euphorbias in the Garden – Good Doers in Different Conditions and Situations Don Witton
©Don Witton ©Don Witton Euphorbias in the garden – good doers in different conditions and situations Don Witton Euphorbias on Don’s allotment in May ith about 2,200 the cold, hard winters of Wknown species, genus central Asia. The hardy leafy euphorbias can be found Euphorbia is the second largest spurges, which Hardy Planters growing in a wide range genus of plants on the planet are more interested in, come of habitats including full, with wild species growing from temperate regions of hot sun through to quite on every continent except Europe and Asia including the deep shade, free-draining Antarctica. Having such a Himalaya region. wide distribution, euphorbias scree to moist, heavier soils. have evolved to survive in There are species which are a wide range of climatic Even within this smaller just a few centimetres high conditions from hot, dry, arid group (around 150 wild to 2.4m+ giants and every parts of Africa and India to species and their forms) size in between. sub-tropical rainforests and ©Don Witton ©Don Witton ©Don Witton Fig.1 E. amygdaloides var. robbiae in the winter garden at Harlow Carr Fig. 2 E. amygdaloides ‘Purpurea’ at Ripley Castle in May 41 ©Don Witton ©Don Witton ©Don Witton Figs 3a & b E. Redwing – a young plant in February & in full bloom E. ‘Blue Haze’ will continue exception is E. characias from Different varieties will grow to flower deep into the the Mediterranean. It is very in a wide range of positions autumn, displaying that variable in the wild and there in the garden and, with wonderfully fresh, chartreuse are 3 subspecies; consequently careful choice, just about acid-yellow colour which there are about 30 named every garden situation can associates so well with many cultivars listed in the Plant other plants. -

Gender Variation in CROTON CALIFORNICUS (EUPHORBIACEAE)
Loma Linda University TheScholarsRepository@LLU: Digital Archive of Research, Scholarship & Creative Works Loma Linda University Electronic Theses, Dissertations & Projects 9-1999 Gender Variation in CROTON CALIFORNICUS (EUPHORBIACEAE) James Lynwood Smith II Follow this and additional works at: https://scholarsrepository.llu.edu/etd Part of the Biology Commons, and the Botany Commons Recommended Citation Smith, James Lynwood II, "Gender Variation in CROTON CALIFORNICUS (EUPHORBIACEAE)" (1999). Loma Linda University Electronic Theses, Dissertations & Projects. 946. https://scholarsrepository.llu.edu/etd/946 This Dissertation is brought to you for free and open access by TheScholarsRepository@LLU: Digital Archive of Research, Scholarship & Creative Works. It has been accepted for inclusion in Loma Linda University Electronic Theses, Dissertations & Projects by an authorized administrator of TheScholarsRepository@LLU: Digital Archive of Research, Scholarship & Creative Works. For more information, please contact [email protected]. LOMA LINDA UNIVERSITY Graduate School GENDER VARIATION IN CROTON CALIFORNICUS (EUPHORBIACEAE) by James Lynwood Smith II A Dissertation in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in Biology September 1999 Each person whose signature appears below certifies that this dissertation in their opinion is adequate, in scope and quality, as a dissertation for the degree of Doctor of Philosophy. , Co-Chairperson , Co-Chairperson Gary L. Bradley, Professor at La Sierra University ii Acknowledgments I wish to thank Ron Carter and Brad Martin for their guidance, assistance, and comments. I am grateful to the other members of my guidance committee, Gary Bradley, Bob Cushman, and Bill Hayes, for their advice and comments. I am also grateful to Aida Smith for assisting with data collection, data entry, and providing comments. -

Survey of Euphorbiaceae Family in Kopergaon Tehsil Of
International Journal of Humanities and Social Sciences (IJHSS) ISSN (P): 2319–393X; ISSN (E): 2319–3948 Vol. 9, Issue 3, Apr–May 2020; 47–58 © IASET SURVEY OF EUPHORBIACEAE FAMILY IN KOPERGAONTEHSIL OF MAHARASHTRA Rahul Chine 1 & MukulBarwant 2 1Research Scholar, Department of Botany, Shri Sadguru Gangagir Maharaj Science College, Maharashtra, India 2Research Scholar, Department of Botany, Sanjivani Arts Commerce and Science College, Maharashtra, India ABSTRACT The survey of Family Euphorbiaceae from Kopargoantehshil is done. In this we first collection of different member of Family Euphorbiaceae from different region of Kopargoantehasil. An extensive and intensive survey at plants was carried out from village Pathare, Derde, Pohegoan, Kopergaon, Padhegaon, Apegoan during the were collected in flowering and fruiting period throughout the year done. During survey we determine 16 member of Euphorbiceae from Kopargoantehshil Then we decide characterization on the basis of habit, flowering character, leaf and fruit character with help of that character and using different literature we identified each and every member of Euphorbiaceae Species were identified with relevant information and documented in this paper with regard to their Botanical Name, family, Habitat, flowering Fruiting session and their medicinal value of some member of Euphorbiaceae that used in medicine respiratory disorder such as cough, asthama, bronchitis etc and some are toxic in nature due to their toxic latex that is showing itching reaction. KEYWORDS : Family Euphorbiaceae, Respiratory Ailment, Identification, Chracterization and Documentation Article History Received: 09 Apr 2020 | Revised: 10 Apr 2020 | Accepted: 18 Apr 2020 INTRODUCTION The Euphorbiaceae, the spurge family, is one of the complex large family of flowering plants of angiosperm with 334 genera and 8000 species in the worlds (Wurdack 2004). -

Myrtle Spurge: Options for Control the Myrtle Spurge, a Class-B Non Desig- the Other Is on Hwy
Myrtle Spurge: Options for Control The myrtle spurge, a class-B non desig- the other is on Hwy. 28 in Odessa. nate noxious weed in Lincoln County, Wash- Myrtle spurge is poisonous if ingested, ington (Euphorbia myrsinites), also known as causing nausea, vomiting and diarrhea. This creeping spurge or donkey tail, and is a suc- plant exudes toxic, milky latex, which can cause culent species of spurges (family Eu- severe skin and eye irritations. Wearing phorbiaceae). Introduced here from gloves, long sleeves, and shoes is highly the Mediterranean region, it is a per- recommended when in contact with ennial forb. It prefers full sun, well Myrtle spurge, as all plant parts are con- drained soil and is found in gardens, sidered poisonous. Although some- natural areas and rocky slopes. Myr- times grown as a decorative plant in tle spurge was added to the Lincoln xeric gardens, myrtle spurge is consid- County ered highly invasive and noxious. This Noxious Weed List plant can rapidly ex- in 2006, after being pand into sensitive eco- discovered in two systems, displacing na- locations. One at tive vegetation and re- Rantz Marina on ducing forage for wild- Lake Roosevelt and life. Key identifying traits • Inconspicuous yellow-green flowers are sur- A close-up of the Myrtle Spurge heart shaped bracts. rounded by heart shaped bracts. Myrtle Spurge is commonly • Plants can grow 8-12 inches tall on ascending found in rock gardens. to trailing stems rising at the tips. • Oval, blue-green, fleshy, succulent-like leaves are arranged in close spirals around the stems. • Stems grow from a prostrate woody base. -

A Survey on Floristic Diversity of Euphorbiaceae Family from Sambhal District of Uttar Pradesh
JASC: Journal of Applied Science and Computations ISSN NO: 1076-5131 A Survey on Floristic Diversity of Euphorbiaceae family from Sambhal district of Uttar Pradesh Santosh Singh Yadav,*Duresh Chand and Zafar Abbas Department of Botany, Gandhi Faiz-E-Aam College , Shahjahanpur -242001,Uttar Pradesh, India Government Degree College,Babrala-Gunnor, Sambhal (U.P.) ABSTRACT There is no detailed information about Angiosperms especially members of family Euphorbiaceae of sambhal district (U.P) .These plants are very important for food value , fodder , biofuel , medicinal and of ornamental value etc . Some of the plants members identified and noted in the study area by monthly trips during 2016-2017 were in all , 18 species under 10 genera which were recorded . These species of Euphorbiaceae were Acalypha indica L (Khokali) , Acalypha wilkesiana Muell , Croton bonplandianum Baill , Emblica officinalis Gaertn (Amla) , Euphorbia milii Ch.des , Euphorbia pulcherrima Willd. , Euphorbia hirta L (Badi dudhi) , Euphorbia thymifolia L (Dudhi) , Jatropha gossypifolia L (Ratanjot wild) , Jatropha curcas Jame ( ratanjot) , Manihot esculenta L. , Pedilanthus tithymaloides L. , Putranjiva roxburghii Wall , Phyllanthus niruri , Ricinus communis Linn , Euphorbia caducifolia Haines (Thor) , Euphorbia granulata Forssk , Phyllanthus reticulatus . This shows economic importance , great diversity and richness in the family Euphorbiaceae of the selected area under study. Geographically , Sambhal (U.P) district coordinates are (28.58 oN and 78.55 oE) it falls under Ramganga and Gangetic plain of North West India. The district of Sambhal has a very rich flora exhibiting diversity of flowering plants. The data collected includes habit, leaf types, fruit types and flowering and fruiting period in the above members of the family Euphorbiaceae in addition to their formal identifications. -

A REVIEW on MEDICINAL USES of DIFFERENT PLANTS of EUPHORBIACEAE FAMILY Md
Shahidul et al. Universal Journal of Pharmaceutical Research 2019; 4(1):45-49 Available online on 15.3.2019 at http://ujpr.org Universal Journal of Pharmaceutical Research An International Peer Reviewed Journal Open access to Pharmaceutical research This is an open access article distributed under the terms of the Creative Commons Attribution-Non Commercial Share Alike 4.0 License which permits unrestricted non commercial use, provided the original work is properly cited Volume 4, Issue 1, 2019 REVIEW ARTICLE A REVIEW ON MEDICINAL USES OF DIFFERENT PLANTS OF EUPHORBIACEAE FAMILY Md. Shahidul Islam*1 , Hasnat Ara1, Kazi Ishtiaq Ahmad2, Md. Mayin Uddin2 1Department of Pharmacy, University of science and Technology Chittagong (USTC), Foy’s Lake, Chittagong, Bangladesh. 2Department of Pharmacy, East West University, Dhaka, Bangladesh. ABSTRACT Euphorbiaceae is an important family which contains numerous medicinal plants. Most of the people in developing countries still today, relays on traditional medicine based largely on species of plants in human being and animals for their primary healthcare. The family Euphorbiaceae is one of the largest family of flowering plants comprising of plants with over 300 genera and 8,000 species. Acalyphaindica L, Euphorbia hirta L, Euphorbia thymifolia L, Croton bonplandianumbaill, Jatropha gossypifolia L, Ricinus communis L are important plants of this family because these plants have different compounds like alkaloids, flavonoids, steroids, saponin, phenolic compounds, fatty acid, esters, minerals etc that have showed different activities in human being and animal. This study provides important data for identification of different plants in Euphorbiaceae family. Species of Euphorbiaceae are extensively used as remedies against several diseases and complaints such as cancer, diabetes, diarrhoea, heart diseases, hemorrhages, hepatitis, jaundice, malaria, ophthalmic diseases, rheumatism and scabies etc. -

Synopsis of Euphorbia (Euphorbiaceae) in the State of São Paulo, Brazil
Phytotaxa 181 (4): 193–215 ISSN 1179-3155 (print edition) www.mapress.com/phytotaxa/ PHYTOTAXA Copyright © 2014 Magnolia Press Article ISSN 1179-3163 (online edition) http://dx.doi.org/10.11646/phytotaxa.181.4.1 Synopsis of Euphorbia (Euphorbiaceae) in the state of São Paulo, Brazil OTÁVIO LUIS MARQUES DA SILVA1,3, INÊS CORDEIRO1 & MARIA BEATRIZ ROSSI CARUZO2 ¹Instituto de Botânica, Secretaria do Meio Ambiente, Cx. Postal 3005, 01061-970, São Paulo, SP, Brazil ²Departamento de Ciências Exatas e da Terra, Universidade Federal de São Paulo, Diadema, SP, Brazil 3Author for correspondence. Email: [email protected] Abstract Euphorbia is the largest genus of Euphorbiaceae and is among the giant genera of Angiosperms. In the state of São Paulo, the genus is represented by 23 species occurring in savannas, high altitude fields, and anthropic areas. This work includes an identification key, photographs, and comments on morphology, habitat, and geographical distribution. We reestablish Euphorbia chrysophylla and recognize Leptopus brasiliensis as a synonym of Euphorbia sciadophila. Six new records for the state of São Paulo are presented: Euphorbia adenoptera, E. bahiensis, E. chrysophylla, E. cordeiroae, E. foliolosa and E. ophthalmica. Eight lectotypes are designated. Key words: Neotropical flora, nomenclatural notes, taxonomy Resumo Euphorbia é o maior gênero de Euphorbiaceae e está entre os maiores de Angiospermas. No Estado de São Paulo, está rep- resentado por 23 espécies ocorrendo no cerrado, campos de altitude e áreas antrópicas. Este trabalho inclui uma chave de identificação, comentários sobre morfologia, habitat e distribuição geográfica. Reestabelecemos Euphorbia chrysophylla e reconhecemos Leptopus brasiliensis como sinônimo de Euphorbia sciadophila. Seis novas ocorrências para o Estado de São Paulo são apresentadas: Euphorbia adenoptera, E. -

Petition for the Release of Aphthona Cyparissiae Against Leafy Spurge in the United States1
Reprinted with permission from: USDA-ARS, March 19, 1986, unpublished. Petition for the release of Aphthona cyparissiae against leafy spurge in the 1 United States ROBERT W. PEMBERTON Part I TO: Dr. R. Bovey, Dept. of Range Science USDA/ARS/SR Texas A&M University College Station, TX 77843 U.S.A. Enclosed are the results of the research on the flea beetle Aphthona cyparissiae (Chrysomelidae). This petition is composed of two parts. The first is the report “Aph- thona cyparissiae (Kock) and A. flava Guill. (Coleopera: Chrysomelidae): Two candi- dates for the biological control of cypress and leafy spurge in North America” by G. Sommer and E. Maw which the Working Group reviewed on Canada’s behalf. Copies of this report should be in the Working Group’s files. This research, which was done at the Commonwealth Institute of Biological Control’s Delémont, Switzerland lab, showed A. cyparissiae to be specific to the genus Euphorbia of the Euphorbiaceae. This result agrees with the literature and field records for A. cyparissiae, which recorded it from: Euphorbia cyparissias, E. esula, E. peplus, E. seguieriana, and E. virgata. The Sommer and Maw report also included information on A. cyparissiae’s taxonomic position, life history, laboratory biology, mortality factors, feeding effects on the host plants, the Harris scoring system, as well as a brief description of the target plant – leafy spurge (Euphorbia esula complex), a serious pest of the rangelands of the Great Plains of North America. Based on this research, the Working Group gave permission to release A. cyparissiae in Canada and to import it into the USDA’s Biological Control of Weeds Quarantine in Al- bany, California, for additional testing. -
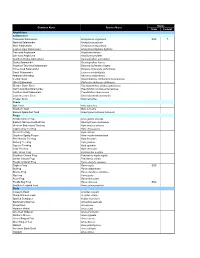
St. Joseph Bay Native Species List
Status Common Name Species Name State Federal Amphibians Salamanders Flatwoods Salamander Ambystoma cingulatum SSC T Marbled Salamander Ambystoma opacum Mole Salamander Ambystoma talpoideum Eastern Tiger Salamander Ambystoma tigrinum tigrinum Two-toed Amphiuma Amphiuma means One-toed Amphiuma Amphiuma pholeter Southern Dusky Salamander Desmognathus auriculatus Dusky Salamander Desmognathus fuscus Southern Two-lined Salamander Eurycea bislineata cirrigera Three-lined Salamander Eurycea longicauda guttolineata Dwarf Salamander Eurycea quadridigitata Alabama Waterdog Necturus alabamensis Central Newt Notophthalmus viridescens louisianensis Slimy Salamander Plethodon glutinosus glutinosus Slender Dwarf Siren Pseudobranchus striatus spheniscus Gulf Coast Mud Salamander Pseudotriton montanus flavissimus Southern Red Salamander Pseudotriton ruber vioscai Eastern Lesser Siren Siren intermedia intermedia Greater Siren Siren lacertina Toads Oak Toad Bufo quercicus Southern Toad Bufo terrestris Eastern Spadefoot Toad Scaphiopus holbrooki holbrooki Frogs Florida Cricket Frog Acris gryllus dorsalis Eastern Narrow-mouthed Frog Gastrophryne carolinensis Western Bird-voiced Treefrog Hyla avivoca avivoca Cope's Gray Treefrog Hyla chrysoscelis Green Treefrog Hyla cinerea Southern Spring Peeper Hyla crucifer bartramiana Pine Woods Treefrog Hyla femoralis Barking Treefrog Hyla gratiosa Squirrel Treefrog Hyla squirella Gray Treefrog Hyla versicolor Little Grass Frog Limnaoedus ocularis Southern Chorus Frog Pseudacris nigrita nigrita Ornate Chorus Frog Pseudacris -

Plant Pest Risk Assessment for Oblong Spurge, Euphorbia Oblongata 2008 (Revised 2013)
Oregon Department of Agriculture Plant Pest Risk Assessment for Oblong Spurge, Euphorbia oblongata 2008 (Revised 2013) Names: Oblong spurge, Euphorbia oblongata, a.k.a. eggleaf spurge Family: Spurge, Euphorbiaceae Findings of This Review and Assessment: Euphorbia oblongata, has been determined to be a category “A” rated noxious weed as defined by the Oregon Department of Agriculture (ODA) Noxious Weed Policy and Classification System. This determination was based on a literature review and analysis using two ODA evaluation forms. Using the Noxious Qualitative Weed Risk Assessment v.3.8, oblong spurge scored 58 indicating a Risk Category of “A”; and a score of 18 with the Noxious Weed Rating System v.3.2, indicating an “A” rating. Oblong spurge scored a very high “A” ranking. Introduction: Oblong spurge is a weedy escaped ornamental species of Euphorbia known from only one site in Salem and one ornamental planting in Eugene, Oregon. Suspected to have been introduced from California in contaminated flax or machinery that was used at the State Penitentiary flax mill in the early part of the 1900’s, it has slowly expanded its territory on the penitentiary property. Growing up to 3’ tall, this species is capable of forming dense stands in more arid climates and could be expected to be a troublesome weed to control should it spread and establish in eastern Oregon. Oblong spurge has a great capacity to infest riparian areas in Washington and Oregon. It is well adapted to a wide range of range, shrub and pine forest environments. In California, it has been found associated with French broom, blackberry and in dry drainages along roadsides. -

Integrated Noxious Weed Management Plan: US Air Force Academy and Farish Recreation Area, El Paso County, CO
Integrated Noxious Weed Management Plan US Air Force Academy and Farish Recreation Area August 2015 CNHP’s mission is to preserve the natural diversity of life by contributing the essential scientific foundation that leads to lasting conservation of Colorado's biological wealth. Colorado Natural Heritage Program Warner College of Natural Resources Colorado State University 1475 Campus Delivery Fort Collins, CO 80523 (970) 491-7331 Report Prepared for: United States Air Force Academy Department of Natural Resources Recommended Citation: Smith, P., S. S. Panjabi, and J. Handwerk. 2015. Integrated Noxious Weed Management Plan: US Air Force Academy and Farish Recreation Area, El Paso County, CO. Colorado Natural Heritage Program, Colorado State University, Fort Collins, Colorado. Front Cover: Documenting weeds at the US Air Force Academy. Photos courtesy of the Colorado Natural Heritage Program © Integrated Noxious Weed Management Plan US Air Force Academy and Farish Recreation Area El Paso County, CO Pam Smith, Susan Spackman Panjabi, and Jill Handwerk Colorado Natural Heritage Program Warner College of Natural Resources Colorado State University Fort Collins, Colorado 80523 August 2015 EXECUTIVE SUMMARY Various federal, state, and local laws, ordinances, orders, and policies require land managers to control noxious weeds. The purpose of this plan is to provide a guide to manage, in the most efficient and effective manner, the noxious weeds on the US Air Force Academy (Academy) and Farish Recreation Area (Farish) over the next 10 years (through 2025), in accordance with their respective integrated natural resources management plans. This plan pertains to the “natural” portions of the Academy and excludes highly developed areas, such as around buildings, recreation fields, and lawns. -

Macbridea Alba
Macbridea alba (White birds-in-a-nest) 5-Year Review: Summary and Evaluation Lathrop Management Area, Bay County. Photos by Vivian Negrón-Ortiz U.S. Fish and Wildlife Service Southeast Region Panama City Field Office Panama City, Florida 5-YEAR REVIEW Macbridea alba (White birds-in-a-nest) I. GENERAL INFORMATION A. Methodology used to complete the review This review was accomplished using information obtained from the plant’s 1994 Recovery Plan, peer reviewed scientific publications, unpublished field survey results, reports of current research projects, unpublished field observations by Service, State and other experienced biologists, and personal communications. These documents are on file at the Panama City Field Office. A Federal Register notice announcing the review and requesting information was published on April 16, 2008 (73 FR 20702). Comments received and suggestions from peer reviewers were evaluated and incorporated as appropriate (see appendix A). No part of this review was contracted to an outside party. This review was completed by the Service’s lead Recovery botanist in the Panama City Field Office, Florida. B. Reviewers Lead Field Office: Dr. Vivian Negrón-Ortiz, Panama City Field Office, 850-769-0552 ext. 231, [email protected] Lead Region: Southeast Region: Kelly Bibb, 404-679-7132 Peer reviewers: Ms. Louise Kirn, District Ecologist Apalachicola National Forest P.O. Box 579, Bristol, FL 32321 Ms. Faye Winters, Field Office Biologist BLM Jackson Field Office 411 Briarwood Drive, Suite 404 Jackson, MS 39206 C. Background 1. FR Notice citation announcing initiation of this review: 73 FR 20702 (April 16, 2008). 1 2. Species status: Unknown (Recovery Data Call 2008); the species status is unknown until all the Element Occurrences1 (EO’s) are revisited.