Genetic, Structural and Clinical Analysis of Spastic Paraplegia 4
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Clinical Spectrum and Genetic Landscape for Hereditary Spastic
Dong et al. Molecular Neurodegeneration (2018) 13:36 https://doi.org/10.1186/s13024-018-0269-1 RESEARCH ARTICLE Open Access Clinical spectrum and genetic landscape for hereditary spastic paraplegias in China En-Lin Dong1†, Chong Wang1†, Shuang Wu1†, Ying-Qian Lu1, Xiao-Hong Lin1, Hui-Zhen Su1, Miao Zhao1, Jin He1, Li-Xiang Ma2, Ning Wang1,3, Wan-Jin Chen1,3* and Xiang Lin1* Abstract Background: Hereditary spastic paraplegias (HSP) is a heterogeneous group of rare neurodegenerative disorders affecting the corticospinal tracts. To date, more than 78 HSP loci have been mapped to cause HSP. However, both the clinical and mutational spectrum of Chinese patients with HSP remained unclear. In this study, we aim to perform a comprehensive analysis of clinical phenotypes and genetic distributions in a large cohort of Chinese HSP patients, and to elucidate the primary pathogenesis in this population. Methods: We firstly performed next-generation sequencing targeting 149 genes correlated with HSP in 99 index cases of our cohort. Multiplex ligation-dependent probe amplification testing was further carried out among those patients without known disease-causing gene mutations. We simultaneously performed a retrospective study on the reported patients exhibiting HSP in other Chinese cohorts. All clinical and molecular characterization from above two groups of Chinese HSP patients were analyzed and summarized. Eventually, we further validated the cellular changes in fibroblasts of two major spastic paraplegia (SPG) patients (SPG4 and SPG11) in vitro. Results: Most patients of ADHSP (94%) are pure forms, whereas most patients of ARHSP (78%) tend to be complicated forms. In ADHSP, we found that SPG4 (79%) was the most prevalent, followed by SPG3A (11%), SPG6 (4%) and SPG33 (2%). -

Identification of the Drosophila Melanogaster Homolog of the Human Spastin Gene
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by RERO DOC Digital Library Dev Genes Evol (2003) 213:412–415 DOI 10.1007/s00427-003-0340-x EXPRESSION NOTE Lars Kammermeier · Jrg Spring · Michael Stierwald · Jean-Marc Burgunder · Heinrich Reichert Identification of the Drosophila melanogaster homolog of the human spastin gene Received: 14 April 2003 / Accepted: 5 May 2003 / Published online: 5 June 2003 Springer-Verlag 2003 Abstract The human SPG4 locus encodes the spastin gene encoding spastin. This gene is expressed ubiqui- gene, which is responsible for the most prevalent form tously in fetal and adult human tissues (Hazan et al. of autosomal dominant hereditary spastic paraplegia 1999). The highest expression levels are found in the (AD-HSP), a neurodegenerative disorder. Here we iden- brain, with selective expression in the cortex and striatum. tify the predicted gene product CG5977 as the Drosophila In the spinal cord, spastin is expressed exclusively in homolog of the human spastin gene, with much higher nuclei of motor neurons, suggesting that the strong sequence similarities than any other related AAA domain neurodegenerative defects observed in patients are caused protein in the fly. Furthermore we report a new potential by a primary defect of spastin in neurons (Charvin et al. transmembrane domain in the N-terminus of the two 2003). The human spastin gene encodes a predicted 616- homologous proteins. During embryogenesis, the expres- amino-acid long protein and is a member of the large sion pattern of Drosophila spastin becomes restricted family of proteins with an AAA domain (ATPases primarily to the central nervous system, in contrast to the Associated with diverse cellular Activities). -

ARCHAEAL EVOLUTION Evolutionary Insights from the Vikings
RESEARCH HIGHLIGHTS Nature Reviews Microbiology | Published online 16 Jan 2017; doi:10.1038/nrmicro.2016.198 ARCHAEAL EVOLUTION Evolutionary insights from the Vikings The emergence of the eukaryotic cell ASGARD — after the invisible the closest known homologue of during evolution gave rise to all com- ‘Gods of Asgard’ in Norse mythology. eukaryotic epsilon DNA polymerases primordial plex life forms on Earth, including The superphylum consists of the identified thus far. eukaryotic multicellular organisms such as ani- previously identified Lokiarchaeota Members of the ASGARD super- mals, plants and fungi. However, the and Thorarchaeota phyla, and the phylum were particularly enriched vesicular and origin of eukaryotes and their char- newly identified Odinarchaeota and for eukaryotic signature proteins trafficking acteristic structural complexity has Heimdallarchaeota phyla. Using that are involved in intracellular components remained a mystery. The most recent phylogenomics, they discovered trafficking and secretion. Several are derived insights into eukaryogenesis support a strong phylogenetic association proteins contained domain signatures the endosymbiotic theory, which between ASGARD lineages and of eukaryotic transport protein from our proposes that the first eukaryotic eukaryotes that placed the eukaryote particle (TRAPP) complexes, which archaeal cell arose from archaea through the lineage in close proximity to the are involved in transport from the ancestor acquisition of an alphaproteobacterial ASGARD superphylum. endoplasmic -

Ubiquitin-Dependent Sorting in Endocytosis
Downloaded from http://cshperspectives.cshlp.org/ on September 29, 2021 - Published by Cold Spring Harbor Laboratory Press Ubiquitin-Dependent Sorting in Endocytosis Robert C. Piper1, Ivan Dikic2, and Gergely L. Lukacs3 1Department of Molecular Physiology and Biophysics, University of Iowa, Iowa City, Iowa 52242 2Buchmann Institute for Molecular Life Sciences and Institute of Biochemistry II, Goethe University School of Medicine, D-60590 Frankfurt, Germany 3Department of Physiology, McGill University, Montre´al, Que´bec H3G 1Y6, Canada Correspondence: [email protected] When ubiquitin (Ub) is attached to membrane proteins on the plasma membrane, it directs them through a series of sorting steps that culminate in their delivery to the lumen of the lysosome where they undergo complete proteolysis. Ubiquitin is recognized by a series of complexes that operate at a number of vesicle transport steps. Ubiquitin serves as a sorting signal for internalization at the plasma membrane and is the major signal for incorporation into intraluminal vesicles of multivesicular late endosomes. The sorting machineries that catalyze these steps can bind Ub via a variety of Ub-binding domains. At the same time, many of these complexes are themselves ubiquitinated, thus providing a plethora of potential mechanisms to regulate their activity. Here we provide an overview of how membrane proteins are selected for ubiquitination and deubiquitination within the endocytic pathway and how that ubiquitin signal is interpreted by endocytic sorting machineries. HOW PROTEINS ARE SELECTED FOR distinctions concerning different types of ligas- UBIQUITINATION es, their specificity for forming particular poly- Ub chains, and even the kinds of residues in biquitin (Ub) is covalently attached to sub- substrate proteins that can be ubiquitin modi- Ustrate proteins by the concerted action of fied, have been blurred with the discovery of E2-conjugating enzymes and E3 ligases (Var- mixed poly-Ub chains, new enzymatic mecha- shavsky 2012). -

Microtubule-Severing Enzymes at the Cutting Edge
Commentary 2561 Microtubule-severing enzymes at the cutting edge David J. Sharp1,* and Jennifer L. Ross2 1Department of Physiology and Biophysics, Albert Einstein College of Medicine, 1300 Morris Park Avenue, Bronx, NY 10461, USA 2Department of Physics, University of Massachusetts-Amherst, 302 Hasbrouck Laboratory, Amherst, MA 01003, USA *Author for correspondence ([email protected]) Journal of Cell Science 125, 2561–2569 ß 2012. Published by The Company of Biologists Ltd doi: 10.1242/jcs.101139 Summary ATP-dependent severing of microtubules was first reported in Xenopus laevis egg extracts in 1991. Two years later this observation led to the purification of the first known microtubule-severing enzyme, katanin. Katanin homologs have now been identified throughout the animal kingdom and in plants. Moreover, members of two closely related enzyme subfamilies, spastin and fidgetin, have been found to sever microtubules and might act alongside katanins in some contexts (Roll-Mecak and McNally, 2010; Yu et al., 2008; Zhang et al., 2007). Over the past few years, it has become clear that microtubule-severing enzymes contribute to a wide range of cellular activities including mitosis and meiosis, morphogenesis, cilia biogenesis and disassembly, and migration. Thus, this group of enzymes is revealing itself to be among the most important of the microtubule regulators. This Commentary focuses on our growing understanding of how microtubule-severing enzymes contribute to the organization and dynamics of diverse microtubule arrays, as well as the structural and biophysical characteristics that afford them the unique capacity to catalyze the removal of tubulin from the interior microtubule lattice. Our goal is to provide a broader perspective, focusing on a limited number of particularly informative, representative and/or timely findings. -

A Deficiency Screen for Genetic Interactions with Spastin Overexpression Reveals a Role for P21-Activated Kinase 3
Genetics:Genetics: PublishedPublished ArticlesArticles AheadAhead ofof Print,Print, publishedpublished onon JuneJuly 29,24, 20112011 asas 10.1534/genetics.111.13083110.1534/genetics.111.130831 1 Loss of Drosophila melanogaster p21-activated kinase 3 (pak3) suppresses defects in synapse structure and function caused by spastin mutations Emily F. Ozdowski*, Sophia Gayle*, Hong Bao§, Bing Zhang§, Nina T. Sherwood* *Department of Biology / Institute for Genome Sciences and Policy Duke University Durham, NC 27710 §Department of Zoology University of Oklahoma Norman, OK 73019 Copyright 2011. E. F. Ozdowski, et al. 2 Running Title: pak3 and spastin in synapse development Keywords: spastin (CG5977) pak3 (CG14895) Microtubule Actin Neuromuscular junction Corresponding Author: Nina Tang Sherwood Duke University Medical Center CARL Bldg Box 3577 Durham, NC 27710 Office phone: (919) 684-8658 Lab phone: (919) 668-3656 Fax: (919) 681-9193 Email: [email protected] E. F. Ozdowski, et al. 3 ABSTRACT Microtubules are dynamic structures that must elongate, disassemble, and be cleaved into smaller pieces for proper neuronal development and function. The AAA ATPase Spastin severs microtubules along their lengths and is thought to regulate the balance between long, stable filaments and shorter fragments that seed extension or are transported. In both Drosophila and humans, loss of Spastin function results in reduction of synaptic connections and disabling motor defects. To gain insight into how spastin is regulated, we screened the Drosophila melanogaster genome for deletions that modify a spastin overexpression phenotype, eye size reduction. One suppressor region deleted p21-activated kinase 3 (pak3), which encodes a member of the Pak family of actin-regulatory enzymes, but whose in vivo function is unknown. -

Why Cells and Viruses Cannot Survive Without an ESCRT
cells Review Why Cells and Viruses Cannot Survive without an ESCRT Arianna Calistri * , Alberto Reale, Giorgio Palù and Cristina Parolin Department of Molecular Medicine, University of Padua, 35121 Padua, Italy; [email protected] (A.R.); [email protected] (G.P.); [email protected] (C.P.) * Correspondence: [email protected] Abstract: Intracellular organelles enwrapped in membranes along with a complex network of vesicles trafficking in, out and inside the cellular environment are one of the main features of eukaryotic cells. Given their central role in cell life, compartmentalization and mechanisms allowing their maintenance despite continuous crosstalk among different organelles have been deeply investigated over the past years. Here, we review the multiple functions exerted by the endosomal sorting complex required for transport (ESCRT) machinery in driving membrane remodeling and fission, as well as in repairing physiological and pathological membrane damages. In this way, ESCRT machinery enables different fundamental cellular processes, such as cell cytokinesis, biogenesis of organelles and vesicles, maintenance of nuclear–cytoplasmic compartmentalization, endolysosomal activity. Furthermore, we discuss some examples of how viruses, as obligate intracellular parasites, have evolved to hijack the ESCRT machinery or part of it to execute/optimize their replication cycle/infection. A special emphasis is given to the herpes simplex virus type 1 (HSV-1) interaction with the ESCRT proteins, considering the peculiarities of this interplay and the need for HSV-1 to cross both the nuclear-cytoplasmic and the cytoplasmic-extracellular environment compartmentalization to egress from infected cells. Citation: Calistri, A.; Reale, A.; Palù, Keywords: ESCRT; viruses; cellular membranes; extracellular vesicles; HSV-1 G.; Parolin, C. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

APP) Traffics from the Cell PNAS PLUS Surface Via Endosomes for Amyloid Β (Aβ) Production in the Trans-Golgi Network
Amyloid precursor protein (APP) traffics from the cell PNAS PLUS surface via endosomes for amyloid β (Aβ) production in the trans-Golgi network Regina Wai-Yan Choya,b, Zhiliang Chenga,b, and Randy Schekmana,b,1 aDepartment of Molecular and Cell Biology and bHoward Hughes Medical Institute, University of California, Berkeley, CA 94720 Contributed by Randy Schekman, May 23, 2012 (sent for review January 14, 2012) Amyloid precursor protein (APP) is processed sequentially by the predominantly in early endosomes (12, 13). Hence, converging β-site APP cleaving enzyme and γ-secretase to generate amyloid β evidence confirmed the importance of the endocytic pathway in (Aβ) peptides, one of the hallmarks of Alzheimer’s disease. The the regulation of Aβ production. To date, there has been limited intracellular location of Aβ production—endosomes or the trans- detailed analysis dissecting the different downstream steps fol- Golgi network (TGN)—remains uncertain. We investigated the role lowing endocytosis to reveal the actual location in which Aβ is of different postendocytic trafficking events in Aβ40 production produced. Potential sites include early or late endosomes or the using an RNAi approach. Depletion of Hrs and Tsg101, acting early multivesicular body (MVB) involved in sorting transmembrane in the multivesicular body pathway, retained APP in early endo- proteins for degradation in the lysosome, as well as along the somes and reduced Aβ40 production. Conversely, depletion of retrograde pathway that mediates the recycling of endosomal CHMP6 and VPS4, acting late in the pathway, rerouted endosomal proteins to the TGN. APP to the TGN for enhanced APP processing. We found that We investigated the importance of different postendocytic VPS35 (retromer)-mediated APP recycling to the TGN was required trafficking steps in the production of Aβ40 by using an RNAi for efficient Aβ40 production. -

Burden of Rare Variants in ALS and Axonal Hereditary Neuropathy Genes Influence Survival In
International Journal of Molecular Sciences Article Burden of Rare Variants in ALS and Axonal Hereditary Neuropathy Genes Influence Survival in ALS: Insights from a Next Generation Sequencing Study of an Italian ALS Cohort Stefania Scarlino 1, Teuta Domi 1, Laura Pozzi 1 , Alessandro Romano 1, Giovanni Battista Pipitone 2, Yuri Matteo Falzone 1,3, Lorena Mosca 4, Silvana Penco 4, Christian Lunetta 5, Valeria Sansone 5,6, Lucio Tremolizzo 7, Raffaella Fazio 3, Federica Agosta 8, 3,8,9,10 2 1,3, , 1, Massimo Filippi , Paola Carrera , Nilo Riva * y and Angelo Quattrini y 1 Experimental Neuropathology Unit, Institute of Experimental Neurology (INSPE), Division of Neuroscience, San Raffaele Scientific Institute, 20132 Milan, Italy; [email protected] (S.S.); [email protected] (T.D.); [email protected] (L.P.); [email protected] (A.R.); [email protected] (Y.M.F.); [email protected] (A.Q.) 2 Laboratory of Clinical Molecular Biology, Unit of Genomics for Human Disease Diagnosis, Division of Genetics and Cell Biology, San Raffaele Scientific Institute, 20132 Milan, Italy; [email protected] (G.B.P.); [email protected] (P.C.) 3 Neurology Unit, San Raffaele Scientific Institute, 20132 Milan, Italy; fazio.raff[email protected] (R.F.); fi[email protected] (M.F.) 4 Medical Genetic Unit, Department of Laboratory Medicine, Niguarda Hospital, 20132 Milan, Italy; [email protected] (L.M.); [email protected] (S.P.) 5 NEuroMuscular Omnicentre (NEMO), Fondazione Serena Onlus, Milan 20132, Italy; [email protected] -
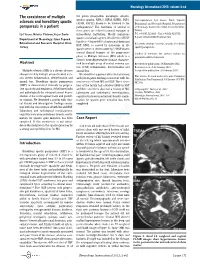
The Coexistence of Multiple Sclerosis and Hereditary Spastic Paraparesis
Neurology International 2013; volume 5:e6 The coexistence of multiple the genes (maspardin, paraplegin, atlastin, spastin, spartin, NIPA 1, KIF5A, HSPD1, PLP1, Correspondence: I ıl Yazıcı, zmir Tepecik sclerosis and hereditary spastic LICAM, BSCL2) known to be involved in the Educatıonal and Researchş Hospıtal,İ Department paraparesis in a patient pathogenesis.8 The functions of several of of Neurology, Gazıler Street 468, Yenı ehır/Izmır, these genes are related to axonal transport or Turkey. ş I ıl Yazıcı, Nılufer Yıldırım,Ya ar Zorlu intracellular trafficking. Mostly mutations Tel. +9.0505.5238031 - Fax: +9.0232.4330756. ş E-mail: [email protected] Department of Neurology, Izmırş Tepecık spastin and atlastin genes identified in ADHSP families.3,4 Around 40% of autosomal dominant Educatıonal and Research Hospıtal, Izmir, Key words: multiple sclerosis, spastin, hereditary HSP, SPG4, is caused by mutations in the Turkey spastic paraparesis. spastin gene on chromosome 2p.6,9 HSP shares several clinical features of the progressive Conflict of interests: the authors declare no phase in Multiple Sclerosis (MS) which is a potential conflict of interests. chronic neurodegenerative disease character- Abstract ized by multiple areas of central nervous sys- Received for publication: 30 November 2012. tem (CNS) inflammation, demyelination and Revision received: 22 January 2013. Multiple sclerosis (MS) is a chronic disease axonal loss. Accepted for publication: 26 February 2013. characterized by multiple areas of central nerv- We identified a patient with clinical history This work is licensed under a Creative Commons ous system inflammation, demyelination and and investigation findings consistent with the Attribution NonCommercial 3.0 License (CC BY- axonal loss. -

Actin, Microtubule, Septin and ESCRT Filament Remodeling During Late Steps of Cytokinesis Cyril Addi, Jian Bai, Arnaud Echard
Actin, microtubule, septin and ESCRT filament remodeling during late steps of cytokinesis Cyril Addi, Jian Bai, Arnaud Echard To cite this version: Cyril Addi, Jian Bai, Arnaud Echard. Actin, microtubule, septin and ESCRT filament remodel- ing during late steps of cytokinesis. Current Opinion in Cell Biology, Elsevier, 2018, 50, pp.27-34. 10.1016/j.ceb.2018.01.007. hal-02114062 HAL Id: hal-02114062 https://hal.archives-ouvertes.fr/hal-02114062 Submitted on 29 Apr 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution - NonCommercial| 4.0 International License Actin, microtubule, septin and ESCRT filament remodeling during late steps of cytokinesis Cyril Addi1,2,3,#, Jian Bai1,2,3,# and Arnaud Echard1,2 1 Membrane Traffic and Cell Division Lab, Cell Biology and Infection department Institut Pasteur, 25–28 rue du Dr Roux, 75724 Paris cedex 15, France 2 Centre National de la Recherche Scientifique CNRS UMR3691, 75015 Paris, France 3 Sorbonne Universités, Université Pierre et Marie Curie, Université Paris 06, Institut de formation doctorale, 75252 Paris, France # equal contribution, alphabetical order correspondence: [email protected] 1 ABSTRACT Cytokinesis is the process by which a mother cell is physically cleaved into two daughter cells.