(E442Q) Redistribute the Wild-Type Spastin Into Filamentous Microtubu
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Clinical Spectrum and Genetic Landscape for Hereditary Spastic
Dong et al. Molecular Neurodegeneration (2018) 13:36 https://doi.org/10.1186/s13024-018-0269-1 RESEARCH ARTICLE Open Access Clinical spectrum and genetic landscape for hereditary spastic paraplegias in China En-Lin Dong1†, Chong Wang1†, Shuang Wu1†, Ying-Qian Lu1, Xiao-Hong Lin1, Hui-Zhen Su1, Miao Zhao1, Jin He1, Li-Xiang Ma2, Ning Wang1,3, Wan-Jin Chen1,3* and Xiang Lin1* Abstract Background: Hereditary spastic paraplegias (HSP) is a heterogeneous group of rare neurodegenerative disorders affecting the corticospinal tracts. To date, more than 78 HSP loci have been mapped to cause HSP. However, both the clinical and mutational spectrum of Chinese patients with HSP remained unclear. In this study, we aim to perform a comprehensive analysis of clinical phenotypes and genetic distributions in a large cohort of Chinese HSP patients, and to elucidate the primary pathogenesis in this population. Methods: We firstly performed next-generation sequencing targeting 149 genes correlated with HSP in 99 index cases of our cohort. Multiplex ligation-dependent probe amplification testing was further carried out among those patients without known disease-causing gene mutations. We simultaneously performed a retrospective study on the reported patients exhibiting HSP in other Chinese cohorts. All clinical and molecular characterization from above two groups of Chinese HSP patients were analyzed and summarized. Eventually, we further validated the cellular changes in fibroblasts of two major spastic paraplegia (SPG) patients (SPG4 and SPG11) in vitro. Results: Most patients of ADHSP (94%) are pure forms, whereas most patients of ARHSP (78%) tend to be complicated forms. In ADHSP, we found that SPG4 (79%) was the most prevalent, followed by SPG3A (11%), SPG6 (4%) and SPG33 (2%). -

Identification of the Drosophila Melanogaster Homolog of the Human Spastin Gene
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by RERO DOC Digital Library Dev Genes Evol (2003) 213:412–415 DOI 10.1007/s00427-003-0340-x EXPRESSION NOTE Lars Kammermeier · Jrg Spring · Michael Stierwald · Jean-Marc Burgunder · Heinrich Reichert Identification of the Drosophila melanogaster homolog of the human spastin gene Received: 14 April 2003 / Accepted: 5 May 2003 / Published online: 5 June 2003 Springer-Verlag 2003 Abstract The human SPG4 locus encodes the spastin gene encoding spastin. This gene is expressed ubiqui- gene, which is responsible for the most prevalent form tously in fetal and adult human tissues (Hazan et al. of autosomal dominant hereditary spastic paraplegia 1999). The highest expression levels are found in the (AD-HSP), a neurodegenerative disorder. Here we iden- brain, with selective expression in the cortex and striatum. tify the predicted gene product CG5977 as the Drosophila In the spinal cord, spastin is expressed exclusively in homolog of the human spastin gene, with much higher nuclei of motor neurons, suggesting that the strong sequence similarities than any other related AAA domain neurodegenerative defects observed in patients are caused protein in the fly. Furthermore we report a new potential by a primary defect of spastin in neurons (Charvin et al. transmembrane domain in the N-terminus of the two 2003). The human spastin gene encodes a predicted 616- homologous proteins. During embryogenesis, the expres- amino-acid long protein and is a member of the large sion pattern of Drosophila spastin becomes restricted family of proteins with an AAA domain (ATPases primarily to the central nervous system, in contrast to the Associated with diverse cellular Activities). -

Microtubule-Severing Enzymes at the Cutting Edge
Commentary 2561 Microtubule-severing enzymes at the cutting edge David J. Sharp1,* and Jennifer L. Ross2 1Department of Physiology and Biophysics, Albert Einstein College of Medicine, 1300 Morris Park Avenue, Bronx, NY 10461, USA 2Department of Physics, University of Massachusetts-Amherst, 302 Hasbrouck Laboratory, Amherst, MA 01003, USA *Author for correspondence ([email protected]) Journal of Cell Science 125, 2561–2569 ß 2012. Published by The Company of Biologists Ltd doi: 10.1242/jcs.101139 Summary ATP-dependent severing of microtubules was first reported in Xenopus laevis egg extracts in 1991. Two years later this observation led to the purification of the first known microtubule-severing enzyme, katanin. Katanin homologs have now been identified throughout the animal kingdom and in plants. Moreover, members of two closely related enzyme subfamilies, spastin and fidgetin, have been found to sever microtubules and might act alongside katanins in some contexts (Roll-Mecak and McNally, 2010; Yu et al., 2008; Zhang et al., 2007). Over the past few years, it has become clear that microtubule-severing enzymes contribute to a wide range of cellular activities including mitosis and meiosis, morphogenesis, cilia biogenesis and disassembly, and migration. Thus, this group of enzymes is revealing itself to be among the most important of the microtubule regulators. This Commentary focuses on our growing understanding of how microtubule-severing enzymes contribute to the organization and dynamics of diverse microtubule arrays, as well as the structural and biophysical characteristics that afford them the unique capacity to catalyze the removal of tubulin from the interior microtubule lattice. Our goal is to provide a broader perspective, focusing on a limited number of particularly informative, representative and/or timely findings. -

A Deficiency Screen for Genetic Interactions with Spastin Overexpression Reveals a Role for P21-Activated Kinase 3
Genetics:Genetics: PublishedPublished ArticlesArticles AheadAhead ofof Print,Print, publishedpublished onon JuneJuly 29,24, 20112011 asas 10.1534/genetics.111.13083110.1534/genetics.111.130831 1 Loss of Drosophila melanogaster p21-activated kinase 3 (pak3) suppresses defects in synapse structure and function caused by spastin mutations Emily F. Ozdowski*, Sophia Gayle*, Hong Bao§, Bing Zhang§, Nina T. Sherwood* *Department of Biology / Institute for Genome Sciences and Policy Duke University Durham, NC 27710 §Department of Zoology University of Oklahoma Norman, OK 73019 Copyright 2011. E. F. Ozdowski, et al. 2 Running Title: pak3 and spastin in synapse development Keywords: spastin (CG5977) pak3 (CG14895) Microtubule Actin Neuromuscular junction Corresponding Author: Nina Tang Sherwood Duke University Medical Center CARL Bldg Box 3577 Durham, NC 27710 Office phone: (919) 684-8658 Lab phone: (919) 668-3656 Fax: (919) 681-9193 Email: [email protected] E. F. Ozdowski, et al. 3 ABSTRACT Microtubules are dynamic structures that must elongate, disassemble, and be cleaved into smaller pieces for proper neuronal development and function. The AAA ATPase Spastin severs microtubules along their lengths and is thought to regulate the balance between long, stable filaments and shorter fragments that seed extension or are transported. In both Drosophila and humans, loss of Spastin function results in reduction of synaptic connections and disabling motor defects. To gain insight into how spastin is regulated, we screened the Drosophila melanogaster genome for deletions that modify a spastin overexpression phenotype, eye size reduction. One suppressor region deleted p21-activated kinase 3 (pak3), which encodes a member of the Pak family of actin-regulatory enzymes, but whose in vivo function is unknown. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Genetic, Structural and Clinical Analysis of Spastic Paraplegia 4
medRxiv preprint doi: https://doi.org/10.1101/2021.07.20.21259482; this version posted July 20, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. It is made available under a CC-BY 4.0 International license . Genetic, structural and clinical analysis of spastic paraplegia 4 Parizad Varghaei1,2, MD, Mehrdad A Estiar2,3, MSc, Setareh Ashtiani4, MSc, Simon Veyron5, PhD, Kheireddin Mufti2,3, MSc, Etienne Leveille6, MD, Eric Yu2,3, BSc, Dan Spiegelman, MSc2, Marie-France Rioux7, MD, Grace Yoon8, MD, Mark Tarnopolsky9, MD, PhD, Kym M. Boycott10, MD, Nicolas Dupre11,12, MD, MSc, Oksana Suchowersky4,13, MD, Jean-François Trempe5,PhD, Guy A. Rouleau2,3,14, MD, PhD, Ziv Gan-Or2,3,14, MD, PhD 1. Division of Experimental Medicine, Department of Medicine, McGill University, Montreal, Quebec, Canada 2. The Neuro (Montreal Neurological Institute-Hospital), McGill University, Montreal, Quebec, Canada 3. Department of Human Genetics, McGill University, Montréal, Québec, Canada 4. Alberta Children’s Hospital, Medical Genetics, Calgary, Alberta, Canada 5. Department of Pharmacology & Therapeutics and Centre de Recherche en Biologie Structurale - FRQS, McGill University, Montréal, Canada 6. Faculty of Medicine, McGill University, Montreal, QC, Canada 7. Department of Neurology, Université de Sherbrooke, Sherbrooke, Québec, Canada. 8. Divisions of Neurology and Clinical and Metabolic Genetics, Department of Paediatrics, University of Toronto, The Hospital for Sick Children, Toronto, Ontario, Canada 9. Department of Pediatrics, McMaster University, Hamilton, Ontario, Canada 10. Children’s Hospital of Eastern Ontario Research Institute, University of Ottawa, Ottawa, Ontario, Canada 11. -

Burden of Rare Variants in ALS and Axonal Hereditary Neuropathy Genes Influence Survival In
International Journal of Molecular Sciences Article Burden of Rare Variants in ALS and Axonal Hereditary Neuropathy Genes Influence Survival in ALS: Insights from a Next Generation Sequencing Study of an Italian ALS Cohort Stefania Scarlino 1, Teuta Domi 1, Laura Pozzi 1 , Alessandro Romano 1, Giovanni Battista Pipitone 2, Yuri Matteo Falzone 1,3, Lorena Mosca 4, Silvana Penco 4, Christian Lunetta 5, Valeria Sansone 5,6, Lucio Tremolizzo 7, Raffaella Fazio 3, Federica Agosta 8, 3,8,9,10 2 1,3, , 1, Massimo Filippi , Paola Carrera , Nilo Riva * y and Angelo Quattrini y 1 Experimental Neuropathology Unit, Institute of Experimental Neurology (INSPE), Division of Neuroscience, San Raffaele Scientific Institute, 20132 Milan, Italy; [email protected] (S.S.); [email protected] (T.D.); [email protected] (L.P.); [email protected] (A.R.); [email protected] (Y.M.F.); [email protected] (A.Q.) 2 Laboratory of Clinical Molecular Biology, Unit of Genomics for Human Disease Diagnosis, Division of Genetics and Cell Biology, San Raffaele Scientific Institute, 20132 Milan, Italy; [email protected] (G.B.P.); [email protected] (P.C.) 3 Neurology Unit, San Raffaele Scientific Institute, 20132 Milan, Italy; fazio.raff[email protected] (R.F.); fi[email protected] (M.F.) 4 Medical Genetic Unit, Department of Laboratory Medicine, Niguarda Hospital, 20132 Milan, Italy; [email protected] (L.M.); [email protected] (S.P.) 5 NEuroMuscular Omnicentre (NEMO), Fondazione Serena Onlus, Milan 20132, Italy; [email protected] -
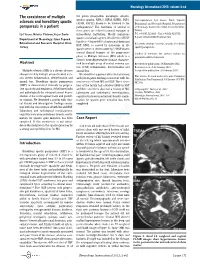
The Coexistence of Multiple Sclerosis and Hereditary Spastic Paraparesis
Neurology International 2013; volume 5:e6 The coexistence of multiple the genes (maspardin, paraplegin, atlastin, spastin, spartin, NIPA 1, KIF5A, HSPD1, PLP1, Correspondence: I ıl Yazıcı, zmir Tepecik sclerosis and hereditary spastic LICAM, BSCL2) known to be involved in the Educatıonal and Researchş Hospıtal,İ Department paraparesis in a patient pathogenesis.8 The functions of several of of Neurology, Gazıler Street 468, Yenı ehır/Izmır, these genes are related to axonal transport or Turkey. ş I ıl Yazıcı, Nılufer Yıldırım,Ya ar Zorlu intracellular trafficking. Mostly mutations Tel. +9.0505.5238031 - Fax: +9.0232.4330756. ş E-mail: [email protected] Department of Neurology, Izmırş Tepecık spastin and atlastin genes identified in ADHSP families.3,4 Around 40% of autosomal dominant Educatıonal and Research Hospıtal, Izmir, Key words: multiple sclerosis, spastin, hereditary HSP, SPG4, is caused by mutations in the Turkey spastic paraparesis. spastin gene on chromosome 2p.6,9 HSP shares several clinical features of the progressive Conflict of interests: the authors declare no phase in Multiple Sclerosis (MS) which is a potential conflict of interests. chronic neurodegenerative disease character- Abstract ized by multiple areas of central nervous sys- Received for publication: 30 November 2012. tem (CNS) inflammation, demyelination and Revision received: 22 January 2013. Multiple sclerosis (MS) is a chronic disease axonal loss. Accepted for publication: 26 February 2013. characterized by multiple areas of central nerv- We identified a patient with clinical history This work is licensed under a Creative Commons ous system inflammation, demyelination and and investigation findings consistent with the Attribution NonCommercial 3.0 License (CC BY- axonal loss. -

Hereditary Spastic Paraplegia: from Genes, Cells and Networks to Novel Pathways for Drug Discovery
brain sciences Review Hereditary Spastic Paraplegia: From Genes, Cells and Networks to Novel Pathways for Drug Discovery Alan Mackay-Sim Griffith Institute for Drug Discovery, Griffith University, Brisbane, QLD 4111, Australia; a.mackay-sim@griffith.edu.au Abstract: Hereditary spastic paraplegia (HSP) is a diverse group of Mendelian genetic disorders affect- ing the upper motor neurons, specifically degeneration of their distal axons in the corticospinal tract. Currently, there are 80 genes or genomic loci (genomic regions for which the causative gene has not been identified) associated with HSP diagnosis. HSP is therefore genetically very heterogeneous. Finding treatments for the HSPs is a daunting task: a rare disease made rarer by so many causative genes and many potential mutations in those genes in individual patients. Personalized medicine through genetic correction may be possible, but impractical as a generalized treatment strategy. The ideal treatments would be small molecules that are effective for people with different causative mutations. This requires identification of disease-associated cell dysfunctions shared across geno- types despite the large number of HSP genes that suggest a wide diversity of molecular and cellular mechanisms. This review highlights the shared dysfunctional phenotypes in patient-derived cells from patients with different causative mutations and uses bioinformatic analyses of the HSP genes to identify novel cell functions as potential targets for future drug treatments for multiple genotypes. Keywords: neurodegeneration; motor neuron disease; spastic paraplegia; endoplasmic reticulum; Citation: Mackay-Sim, A. Hereditary protein-protein interaction network Spastic Paraplegia: From Genes, Cells and Networks to Novel Pathways for Drug Discovery. Brain Sci. 2021, 11, 403. -

Interorganelle Communication, Aging, and Neurodegeneration
Downloaded from genesdev.cshlp.org on October 9, 2021 - Published by Cold Spring Harbor Laboratory Press REVIEW Interorganelle communication, aging, and neurodegeneration Maja Petkovic,1 Caitlin E. O’Brien,2 and Yuh Nung Jan1,2,3 1Department of Physiology, University of California at San Francisco, San Francisco, California 94158, USA; 2Howard Hughes Medical Institute, University of California at San Francisco, San Francisco, California 94158, USA; 3Department of Biochemistry and Biophysics, University of California at San Francisco, San Francisco, California 94158, USA Our cells are comprised of billions of proteins, lipids, and ular transport cannot account for the exchange of certain other small molecules packed into their respective subcel- biomolecules. For example, delivery of several classes of lip- lular organelles, with the daunting task of maintaining cel- ids from their site of synthesis in the endoplasmic reticu- lular homeostasis over a lifetime. However, it is becoming lum (ER) to their steady-state location is unaffected by increasingly evident that organelles do not act as autono- pharmacological and genetic manipulations that deplete mous discrete units but rather as interconnected hubs cellular ATP levels, disrupt vesicular transport, and alter that engage in extensive communication through mem- cytoskeletal dynamics (Kaplan and Simoni 1985; Vance brane contacts. In the last few years, our understanding et al. 1991; Hanada et al. 2003; Baumann et al. 2005; Lev of how these contacts coordinate organelle function has re- 2012). Moreover, intracellular compartments like the ER defined our view of the cell. This review aims to present and mitochondria refill their depleted calcium (Ca2+) stores novel findings on the cellular interorganelle communica- through low-affinity calcium channels that require much tion network and how its dysfunction may contribute to higher local concentrations of calcium than global cytosol- aging and neurodegeneration. -

Identification of Genetic Modifiers in Hereditary Spastic Paraplegias Due to SPAST/SPG4 Mutations Livia Parodi
Identification of genetic modifiers in Hereditary Spastic Paraplegias due to SPAST/SPG4 mutations Livia Parodi To cite this version: Livia Parodi. Identification of genetic modifiers in Hereditary Spastic Paraplegias due to SPAST/SPG4 mutations. Human health and pathology. Sorbonne Université, 2019. English. NNT : 2019SORUS317. tel-03141229 HAL Id: tel-03141229 https://tel.archives-ouvertes.fr/tel-03141229 Submitted on 15 Feb 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Sorbonne Université Institut du Cerveau et de la Moelle Épinière École Doctorale Cerveau-Cognition-Comportement Thèse de doctorat en Neurosciences Identification of genetic modifiers in Hereditary Spastic Paraplegias due to SPAST/SPG4 mutations Soutenue le 9 octobre 2019 par Livia Parodi Membres du jury : Pr Bruno Stankoff Président Pr Lesley Jones Rapporteur Dr Susanne de Bot Rapporteur Pr Christel Depienne Examinateur Pr Cyril Goizet Examinateur Pr Alexandra Durr Directeur de thèse Table of contents Abbreviations _________________________________________________________ -

Truncating Mutations in SPAST Patients Are Associated with a High
JNNP Online First, published on June 1, 2017 as 10.1136/jnnp-2017-315796 J Neurol Neurosurg Psychiatry: first published as 10.1136/jnnp-2017-315796 on 1 June 2017. Downloaded from Neurogenetics RESEARCH PAPER Truncating mutations in SPAST patients are associated with a high rate of psychiatric comorbidities in hereditary spastic paraplegia Viorica Chelban,1,2,3 Arianna Tucci,1,2,4,5 David S Lynch,1,2 James M Polke,1,2,6 Liana Santos,6 Hallgeir Jonvik,1,2 Stanislav Groppa,3 Nicholas W Wood,1,2 Henry Houlden1,2,6 ► Additional material is ABSTRact rarely described in SPAST and include seizures, intel- published online only. To view Background The hereditary spastic paraplegias lectual disability and cerebellar ataxia.11–13 Neuro- please visit the journal online psychiatric comorbidities have only been reported (http:// dx. doi. org/ 10. 1136/ (HSPs) are a rare and heterogeneous group of jnnp- 2017- 315796). neurodegenerative disorders that are clinically in one study, where a higher-than-expected rate characterised by progressive lower limb spasticity. They of psychosis was found in individuals with HSPs 1Department of Molecular are classified as either ‘pure’ or ‘complex’ where spastic including SPAST;14 however, the association with Neuroscience, UCL Institute of paraplegia is complicated with additional neurological spastin was uncertain. Depression is reported to be Neurology, London, UK 15 2National Hospital for Neurology features. Mutations in the spastin gene (SPAST) are the frequent (41%) but unrelated to disease severity. and Neurosurgery, London, UK most common cause of HSP and typically present with a No clear correlation between type or location of the 3Department of Neurology pure form.