Cytotype Variation, Cryptic Diversity and Hybridization in Ranunculus Sect
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Introduction to Common Native & Invasive Freshwater Plants in Alaska
Introduction to Common Native & Potential Invasive Freshwater Plants in Alaska Cover photographs by (top to bottom, left to right): Tara Chestnut/Hannah E. Anderson, Jamie Fenneman, Vanessa Morgan, Dana Visalli, Jamie Fenneman, Lynda K. Moore and Denny Lassuy. Introduction to Common Native & Potential Invasive Freshwater Plants in Alaska This document is based on An Aquatic Plant Identification Manual for Washington’s Freshwater Plants, which was modified with permission from the Washington State Department of Ecology, by the Center for Lakes and Reservoirs at Portland State University for Alaska Department of Fish and Game US Fish & Wildlife Service - Coastal Program US Fish & Wildlife Service - Aquatic Invasive Species Program December 2009 TABLE OF CONTENTS TABLE OF CONTENTS Acknowledgments ............................................................................ x Introduction Overview ............................................................................. xvi How to Use This Manual .................................................... xvi Categories of Special Interest Imperiled, Rare and Uncommon Aquatic Species ..................... xx Indigenous Peoples Use of Aquatic Plants .............................. xxi Invasive Aquatic Plants Impacts ................................................................................. xxi Vectors ................................................................................. xxii Prevention Tips .................................................... xxii Early Detection and Reporting -

Aquatic Vascular Plant Species Distribution Maps
Appendix 11.5.1: Aquatic Vascular Plant Species Distribution Maps These distribution maps are for 116 aquatic vascular macrophyte species (Table 1). Aquatic designation follows habitat descriptions in Haines and Vining (1998), and includes submergent, floating and some emergent species. See Appendix 11.4 for list of species. Also included in Appendix 11.4 is the number of HUC-10 watersheds from which each taxon has been recorded, and the county-level distributions. Data are from nine sources, as compiled in the MABP database (plus a few additional records derived from ancilliary information contained in reports from two fisheries surveys in the Upper St. John basin organized by The Nature Conservancy). With the exception of the University of Maine herbarium records, most locations represent point samples (coordinates were provided in data sources or derived by MABP from site descriptions in data sources). The herbarium data are identified only to township. In the species distribution maps, town-level records are indicated by center-points (centroids). Figure 1 on this page shows as polygons the towns where taxon records are identified only at the town level. Data Sources: MABP ID MABP DataSet Name Provider 7 Rare taxa from MNAP lake plant surveys D. Cameron, MNAP 8 Lake plant surveys D. Cameron, MNAP 35 Acadia National Park plant survey C. Greene et al. 63 Lake plant surveys A. Dieffenbacher-Krall 71 Natural Heritage Database (rare plants) MNAP 91 University of Maine herbarium database C. Campbell 183 Natural Heritage Database (delisted species) MNAP 194 Rapid bioassessment surveys D. Cameron, MNAP 207 Invasive aquatic plant records MDEP Maps are in alphabetical order by species name. -

Predictive Modelling of Spatial Biodiversity Data to Support Ecological Network Mapping: a Case Study in the Fens
Predictive modelling of spatial biodiversity data to support ecological network mapping: a case study in the Fens Christopher J Panter, Paul M Dolman, Hannah L Mossman Final Report: July 2013 Supported and steered by the Fens for the Future partnership and the Environment Agency www.fensforthefuture.org.uk Published by: School of Environmental Sciences, University of East Anglia, Norwich, NR4 7TJ, UK Suggested citation: Panter C.J., Dolman P.M., Mossman, H.L (2013) Predictive modelling of spatial biodiversity data to support ecological network mapping: a case study in the Fens. University of East Anglia, Norwich. ISBN: 978-0-9567812-3-9 © Copyright rests with the authors. Acknowledgements This project was supported and steered by the Fens for the Future partnership. Funding was provided by the Environment Agency (Dominic Coath). We thank all of the species recorders and natural historians, without whom this work would not be possible. Cover picture: Extract of a map showing the predicted distribution of biodiversity. Contents Executive summary .................................................................................................................... 4 Introduction ............................................................................................................................... 5 Methodology .......................................................................................................................... 6 Biological data ................................................................................................................... -

(2009) Red Data List of Derbyshire's Vascular Plants
Red Data List of Derbyshire’s Vascular Plants Moyes, N.J. & Willmot, A. Derby Museum & Art Gallery 2009 Contents 1. Introduction Page 2 2. Red Data List Categories – What’s Included? Page 3 3. What’s Not Included? Page 4 4. Conclusion & Recommendations Page 4 5. Table 1 List of Category 1 Plants Page 5 6. Table 2 List of Category 2 Plants Page 5 7. Table 3 List of Category 3 Plants Page 7 8. Table 4 List of Category 4 Plants Page 8 9. Table 5 List of Category 5 Plants Page 9 10. Table 6 List of Category 6 Plants Page 11 11. References Page 12 Appendix 1 History of Derbyshire Red Data Lists Page 13 Appendix 2 Assessing Local Decline Page 15 Appendix 3 Full List of Derbyshire Red Data Plants Page 18 CITATION: Moyes, N.J. & Willmot, A. (2009) Red Data List of Derbyshire’s Vascular Plants. Derby Museum. 1 1) Introduction County Rare Plant Lists – or Red Data Lists – are a valuable tool to identify species of conservation concern at the local level. These are the plants we should be most concerned about protecting when they are still present, or looking out for if they seem to have declined or become extinct in the locality. All the species named in this Red Data List are native vascular plants in the area, and they either: have a national conservation status in the UK, or are rare in Derbyshire, or have exhibited a significant local decline in recent times, or have become locally extinct. The geographic area in the definition of Derbyshire used here includes: the modern administrative county of Derbyshire, the City of Derby the historic botanical recording area known as the “vice-county” of Derbyshire (VC57). -

Ranunculaceae.Pdf
Botanische Bestimmungsübungen 1 Ranunculaceae Ranunculaceae Hahnenfußgewächse 1 Systematik und Verbreitung Die Ranunculaceae gehören zu den Eudikotyledonen. Innerhalb dieser werden sie zur Ordnung der Ranunculales (Hahnenfußartige) gestellt. Die Ranunculaceae umfassen rund 60 Gattungen mit insgesamt etwa 2500 Arten und werden derzeit in 5 Unterfamilien eingeteilt: 1. Glaucidioideae (z. B. Glaucidium, Scheinhahnenfuß), 2. Hydrastidoideae (z. B. Hydrastis, Gelbwurz), 3. Coptoideae (z. B. Coptis, Gold- faden), 4. Thalictroideae (z. B. Thalictrum, Wiesenraute) und 5. Ranunculoideae (z. B. Ranunculus, Hahnenfuß). Letztere wird in 10 Triben untergliedert Hahnenfußgewächse sind kosmopolitisch verbreitet mit einem Schwerpunkt in den gemäßigten bis kühleren Zonen der Nordhemisphäre. Das Diversitätszentrum liegt in O-Asien. Der Großteil der bei uns heimischen Arten stammt aus der Gattung Ranunculus (Hahnenfuß). Abb. 1: Verbreitungskarte. 2 Morphologie 2.1 Habitus Der Großteil der Arten sind krautige Pflanzen. Wenige Arten, z. B. innerhalb der Gattung Clematis (Waldrebe), sind verholzte Kletterpflanzen. Bei einigen Arten, z. B. Eranthis hyemalis (Winterling), handelt es sich um Geophyten mit einer Knolle © PD DR. VEIT M. DÖRKEN, Universität Konstanz, FB Biologie Botanische Bestimmungsübungen 2 Ranunculaceae als unterirdisches Überdauerungsorgan. Einige Arten, z. B. Ranunculus repens (Kriechender Hahnenfuß), treiben oberirdische Ausläufer. Andere Arten wie Anemone nemorosa (Busch-Windröschen), hingegen haben ein kräftiges unterirdisches Rhizom (Kriechspross). -

THE IRISH RED DATA BOOK 1 Vascular Plants
THE IRISH RED DATA BOOK 1 Vascular Plants T.G.F.Curtis & H.N. McGough Wildlife Service Ireland DUBLIN PUBLISHED BY THE STATIONERY OFFICE 1988 ISBN 0 7076 0032 4 This version of the Red Data Book was scanned from the original book. The original book is A5-format, with 168 pages. Some changes have been made as follows: NOMENCLATURE has been updated, with the name used in the 1988 edition in brackets. Irish Names and family names have also been added. STATUS: There have been three Flora Protection Orders (1980, 1987, 1999) to date. If a species is currently protected (i.e. 1999) this is stated as PROTECTED, if it was previously protected, the year(s) of the relevant orders are given. IUCN categories have been updated as follows: EN to CR, V to EN, R to V. The original (1988) rating is given in brackets thus: “CR (EN)”. This takes account of the fact that a rare plant is not necessarily threatened. The European IUCN rating was given in the original book, here it is changed to the UK IUCN category as given in the 2005 Red Data Book listing. MAPS and APPENDIX have not been reproduced here. ACKNOWLEDGEMENTS We are most grateful to the following for their help in the preparation of the Irish Red Data Book:- Christine Leon, CMC, Kew for writing the Preface to this Red Data Book and for helpful discussions on the European aspects of rare plant conservation; Edwin Wymer, who designed the cover and who, as part of his contract duties in the Wildlife Service, organised the computer applications to the data in an efficient and thorough manner. -
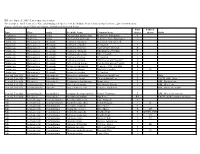
Threatened and Endangered Species List
Effective April 15, 2009 - List is subject to revision For a complete list of Tennessee's Rare and Endangered Species, visit the Natural Areas website at http://tennessee.gov/environment/na/ Aquatic and Semi-aquatic Plants and Aquatic Animals with Protected Status State Federal Type Class Order Scientific Name Common Name Status Status Habit Amphibian Amphibia Anura Gyrinophilus gulolineatus Berry Cave Salamander T Amphibian Amphibia Anura Gyrinophilus palleucus Tennessee Cave Salamander T Crustacean Malacostraca Decapoda Cambarus bouchardi Big South Fork Crayfish E Crustacean Malacostraca Decapoda Cambarus cymatilis A Crayfish E Crustacean Malacostraca Decapoda Cambarus deweesae Valley Flame Crayfish E Crustacean Malacostraca Decapoda Cambarus extraneus Chickamauga Crayfish T Crustacean Malacostraca Decapoda Cambarus obeyensis Obey Crayfish T Crustacean Malacostraca Decapoda Cambarus pristinus A Crayfish E Crustacean Malacostraca Decapoda Cambarus williami "Brawley's Fork Crayfish" E Crustacean Malacostraca Decapoda Fallicambarus hortoni Hatchie Burrowing Crayfish E Crustacean Malocostraca Decapoda Orconectes incomptus Tennessee Cave Crayfish E Crustacean Malocostraca Decapoda Orconectes shoupi Nashville Crayfish E LE Crustacean Malocostraca Decapoda Orconectes wrighti A Crayfish E Fern and Fern Ally Filicopsida Polypodiales Dryopteris carthusiana Spinulose Shield Fern T Bogs Fern and Fern Ally Filicopsida Polypodiales Dryopteris cristata Crested Shield-Fern T FACW, OBL, Bogs Fern and Fern Ally Filicopsida Polypodiales Trichomanes boschianum -

Vascular Plants of Santa Cruz County, California
ANNOTATED CHECKLIST of the VASCULAR PLANTS of SANTA CRUZ COUNTY, CALIFORNIA SECOND EDITION Dylan Neubauer Artwork by Tim Hyland & Maps by Ben Pease CALIFORNIA NATIVE PLANT SOCIETY, SANTA CRUZ COUNTY CHAPTER Copyright © 2013 by Dylan Neubauer All rights reserved. No part of this publication may be reproduced without written permission from the author. Design & Production by Dylan Neubauer Artwork by Tim Hyland Maps by Ben Pease, Pease Press Cartography (peasepress.com) Cover photos (Eschscholzia californica & Big Willow Gulch, Swanton) by Dylan Neubauer California Native Plant Society Santa Cruz County Chapter P.O. Box 1622 Santa Cruz, CA 95061 To order, please go to www.cruzcps.org For other correspondence, write to Dylan Neubauer [email protected] ISBN: 978-0-615-85493-9 Printed on recycled paper by Community Printers, Santa Cruz, CA For Tim Forsell, who appreciates the tiny ones ... Nobody sees a flower, really— it is so small— we haven’t time, and to see takes time, like to have a friend takes time. —GEORGIA O’KEEFFE CONTENTS ~ u Acknowledgments / 1 u Santa Cruz County Map / 2–3 u Introduction / 4 u Checklist Conventions / 8 u Floristic Regions Map / 12 u Checklist Format, Checklist Symbols, & Region Codes / 13 u Checklist Lycophytes / 14 Ferns / 14 Gymnosperms / 15 Nymphaeales / 16 Magnoliids / 16 Ceratophyllales / 16 Eudicots / 16 Monocots / 61 u Appendices 1. Listed Taxa / 76 2. Endemic Taxa / 78 3. Taxa Extirpated in County / 79 4. Taxa Not Currently Recognized / 80 5. Undescribed Taxa / 82 6. Most Invasive Non-native Taxa / 83 7. Rejected Taxa / 84 8. Notes / 86 u References / 152 u Index to Families & Genera / 154 u Floristic Regions Map with USGS Quad Overlay / 166 “True science teaches, above all, to doubt and be ignorant.” —MIGUEL DE UNAMUNO 1 ~ACKNOWLEDGMENTS ~ ANY THANKS TO THE GENEROUS DONORS without whom this publication would not M have been possible—and to the numerous individuals, organizations, insti- tutions, and agencies that so willingly gave of their time and expertise. -

Ranunculus Sect. Batrachium (Ranunculaceae)
ZOBODAT - www.zobodat.at Zoologisch-Botanische Datenbank/Zoological-Botanical Database Digitale Literatur/Digital Literature Zeitschrift/Journal: Neilreichia - Zeitschrift für Pflanzensystematik und Floristik Österreichs Jahr/Year: 2016 Band/Volume: 8 Autor(en)/Author(s): Englmaier Peter F. J. Artikel/Article: Ranunculus sect. Batrachium (Ranunculaceae): Contribution to an excursion flora of Austria and the Eastern Alps 97-125 ©Verein zur Erforschung der Flora Österreichs; download unter www.zobodat.at Neilreichia 8: 97–125 (2016) Ranunculus sect. Batrachium (Ranunculaceae): Contribution to an excursion flora of Austria and the Eastern Alps Peter Englmaier Faculty of Life Sciences, University of Vienna, Althanstraße 14, 1090 Vienna, Austria; e-mail: [email protected] Abstract: A treatment of Ranunculus sect. Batrachium is presented for the Eastern Alpine terri- tory as covered by the forthcoming 4th ed. of the “Excursion flora of Austria and the entire Eastern Alps”. A newly designed key as well as alternative keys for terrestrial modificants and vegetative specimens are presented. A new outline for the section is applied including (1) the Ranunculus fluitans group (R. fluitans and R. pseudofluitans); (2) the R. trichophyllus group (R. trichophyl lus, R. confervoides, and R. rionii); (3) the R. aquatilis group, including the R. aquatilis subgroup with R. aquatilis (the only member present in Europe), the R. peltatus subgroup (R. peltatus and R. baudotii) as well as related hybridogenous taxa (R. penicillatus s. str.) and (4) R. circinatus. The distribution of all species is given for the Austrian federal states, the Bavarian Alps, Liechtenstein and the South Alpine regions of Switzerland, Italy and Slovenia. -

Conservation Genetics of Ireland's Sole Population of the River Water Crowfoot (Ranunculus Fluitans Lam.)
Conservation genetics of Ireland's sole population of the River water crowfoot (Ranunculus fluitans Lam.) Bradley, C., Duignan, C., Preston, S. J., & Provan, J. (2013). Conservation genetics of Ireland's sole population of the River water crowfoot (Ranunculus fluitans Lam.). Aquatic Botany, 107, 54–58. https://doi.org/10.1016/j.aquabot.2013.01.011 Published in: Aquatic Botany Document Version: Peer reviewed version Queen's University Belfast - Research Portal: Link to publication record in Queen's University Belfast Research Portal Publisher rights This is the author’s version of a work that was accepted for publication in Aquatic Botany. Changes resulting from the publishing process, such as peer review, editing, corrections, structural formatting, and other quality control mechanisms may not be reflected in this document. Changes may have been made to this work since it was submitted for publication. A definitive version was subsequently published in Aquatic Botany, [VOL 107, (2013)] General rights Copyright for the publications made accessible via the Queen's University Belfast Research Portal is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The Research Portal is Queen's institutional repository that provides access to Queen's research output. Every effort has been made to ensure that content in the Research Portal does not infringe any person's rights, or applicable UK laws. If you discover content in the Research Portal that you believe breaches copyright or violates any law, please contact [email protected]. -

Waterton Lakes National Park • Common Name(Order Family Genus Species)
Waterton Lakes National Park Flora • Common Name(Order Family Genus species) Monocotyledons • Arrow-grass, Marsh (Najadales Juncaginaceae Triglochin palustris) • Arrow-grass, Seaside (Najadales Juncaginaceae Triglochin maritima) • Arrowhead, Northern (Alismatales Alismataceae Sagittaria cuneata) • Asphodel, Sticky False (Liliales Liliaceae Triantha glutinosa) • Barley, Foxtail (Poales Poaceae/Gramineae Hordeum jubatum) • Bear-grass (Liliales Liliaceae Xerophyllum tenax) • Bentgrass, Alpine (Poales Poaceae/Gramineae Podagrostis humilis) • Bentgrass, Creeping (Poales Poaceae/Gramineae Agrostis stolonifera) • Bentgrass, Green (Poales Poaceae/Gramineae Calamagrostis stricta) • Bentgrass, Spike (Poales Poaceae/Gramineae Agrostis exarata) • Bluegrass, Alpine (Poales Poaceae/Gramineae Poa alpina) • Bluegrass, Annual (Poales Poaceae/Gramineae Poa annua) • Bluegrass, Arctic (Poales Poaceae/Gramineae Poa arctica) • Bluegrass, Plains (Poales Poaceae/Gramineae Poa arida) • Bluegrass, Bulbous (Poales Poaceae/Gramineae Poa bulbosa) • Bluegrass, Canada (Poales Poaceae/Gramineae Poa compressa) • Bluegrass, Cusick's (Poales Poaceae/Gramineae Poa cusickii) • Bluegrass, Fendler's (Poales Poaceae/Gramineae Poa fendleriana) • Bluegrass, Glaucous (Poales Poaceae/Gramineae Poa glauca) • Bluegrass, Inland (Poales Poaceae/Gramineae Poa interior) • Bluegrass, Fowl (Poales Poaceae/Gramineae Poa palustris) • Bluegrass, Patterson's (Poales Poaceae/Gramineae Poa pattersonii) • Bluegrass, Kentucky (Poales Poaceae/Gramineae Poa pratensis) • Bluegrass, Sandberg's (Poales -

Dulin and Kirchoff 2010
Bot. Rev. DOI 10.1007/s12229-010-9057-5 Paedomorphosis, Secondary Woodiness, and Insular Woodiness in Plants Max W. Dulin1 & Bruce K. Kirchoff1,2 1 Department of Biology, University of North Carolina at Greensboro, 321 McIver St., P. O. Box 26170, Greensboro, NC 27402-6170, USA 2 Author for Correspondence; e-mail: [email protected] # The New York Botanical Garden 2010 TABLE OF CONTENTS I. Abstract ................................................................................................................ II. Introduction ......................................................................................................... III. Historical Review............................................................................................... A. Paedomorphosis, Insular Woodiness, and Secondary Woodiness.............. B. Paedomorphosis........................................................................................... 1. Genesis of the Theory of Paedomorphosis......................................... 2. Bailey’s Refugium Theory and Major Trends in Xylem Evolution... 3. Paedomorphic Characters of the Secondary Xylem........................... C. Insular Woodiness....................................................................................... 1. Examples of Insular Woodiness......................................................... 2. Relictual and Secondary Insular Woodiness...................................... 3. Hypotheses to Expain the Evolution of Insular Woodiness............... D. Secondary Woodiness ................................................................................