Generalized Linear Models
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Generalized Linear Models (Glms)
San Jos´eState University Math 261A: Regression Theory & Methods Generalized Linear Models (GLMs) Dr. Guangliang Chen This lecture is based on the following textbook sections: • Chapter 13: 13.1 – 13.3 Outline of this presentation: • What is a GLM? • Logistic regression • Poisson regression Generalized Linear Models (GLMs) What is a GLM? In ordinary linear regression, we assume that the response is a linear function of the regressors plus Gaussian noise: 0 2 y = β0 + β1x1 + ··· + βkxk + ∼ N(x β, σ ) | {z } |{z} linear form x0β N(0,σ2) noise The model can be reformulate in terms of • distribution of the response: y | x ∼ N(µ, σ2), and • dependence of the mean on the predictors: µ = E(y | x) = x0β Dr. Guangliang Chen | Mathematics & Statistics, San Jos´e State University3/24 Generalized Linear Models (GLMs) beta=(1,2) 5 4 3 β0 + β1x b y 2 y 1 0 −1 0.0 0.2 0.4 0.6 0.8 1.0 x x Dr. Guangliang Chen | Mathematics & Statistics, San Jos´e State University4/24 Generalized Linear Models (GLMs) Generalized linear models (GLM) extend linear regression by allowing the response variable to have • a general distribution (with mean µ = E(y | x)) and • a mean that depends on the predictors through a link function g: That is, g(µ) = β0x or equivalently, µ = g−1(β0x) Dr. Guangliang Chen | Mathematics & Statistics, San Jos´e State University5/24 Generalized Linear Models (GLMs) In GLM, the response is typically assumed to have a distribution in the exponential family, which is a large class of probability distributions that have pdfs of the form f(x | θ) = a(x)b(θ) exp(c(θ) · T (x)), including • Normal - ordinary linear regression • Bernoulli - Logistic regression, modeling binary data • Binomial - Multinomial logistic regression, modeling general cate- gorical data • Poisson - Poisson regression, modeling count data • Exponential, Gamma - survival analysis Dr. -

Bayesian Inference Chapter 4: Regression and Hierarchical Models
Bayesian Inference Chapter 4: Regression and Hierarchical Models Conchi Aus´ınand Mike Wiper Department of Statistics Universidad Carlos III de Madrid Master in Business Administration and Quantitative Methods Master in Mathematical Engineering Conchi Aus´ınand Mike Wiper Regression and hierarchical models Masters Programmes 1 / 35 Objective AFM Smith Dennis Lindley We analyze the Bayesian approach to fitting normal and generalized linear models and introduce the Bayesian hierarchical modeling approach. Also, we study the modeling and forecasting of time series. Conchi Aus´ınand Mike Wiper Regression and hierarchical models Masters Programmes 2 / 35 Contents 1 Normal linear models 1.1. ANOVA model 1.2. Simple linear regression model 2 Generalized linear models 3 Hierarchical models 4 Dynamic models Conchi Aus´ınand Mike Wiper Regression and hierarchical models Masters Programmes 3 / 35 Normal linear models A normal linear model is of the following form: y = Xθ + ; 0 where y = (y1;:::; yn) is the observed data, X is a known n × k matrix, called 0 the design matrix, θ = (θ1; : : : ; θk ) is the parameter set and follows a multivariate normal distribution. Usually, it is assumed that: 1 ∼ N 0 ; I : k φ k A simple example of normal linear model is the simple linear regression model T 1 1 ::: 1 where X = and θ = (α; β)T . x1 x2 ::: xn Conchi Aus´ınand Mike Wiper Regression and hierarchical models Masters Programmes 4 / 35 Normal linear models Consider a normal linear model, y = Xθ + . A conjugate prior distribution is a normal-gamma distribution: -
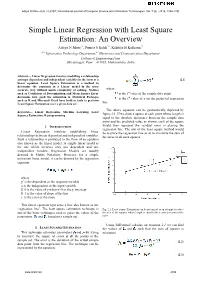
Simple Linear Regression with Least Square Estimation: an Overview
Aditya N More et al, / (IJCSIT) International Journal of Computer Science and Information Technologies, Vol. 7 (6) , 2016, 2394-2396 Simple Linear Regression with Least Square Estimation: An Overview Aditya N More#1, Puneet S Kohli*2, Kshitija H Kulkarni#3 #1-2Information Technology Department,#3 Electronics and Communication Department College of Engineering Pune Shivajinagar, Pune – 411005, Maharashtra, India Abstract— Linear Regression involves modelling a relationship amongst dependent and independent variables in the form of a (2.1) linear equation. Least Square Estimation is a method to determine the constants in a Linear model in the most accurate way without much complexity of solving. Metrics where such as Coefficient of Determination and Mean Square Error is the ith value of the sample data point determine how good the estimation is. Statistical Packages is the ith value of y on the predicted regression such as R and Microsoft Excel have built in tools to perform Least Square Estimation over a given data set. line The above equation can be geometrically depicted by Keywords— Linear Regression, Machine Learning, Least Squares Estimation, R programming figure 2.1. If we draw a square at each point whose length is equal to the absolute difference between the sample data point and the predicted value as shown, each of the square would then represent the residual error in placing the I. INTRODUCTION regression line. The aim of the least square method would Linear Regression involves establishing linear be to place the regression line so as to minimize the sum of relationships between dependent and independent variables. -

Bayesian Methods: Review of Generalized Linear Models
Bayesian Methods: Review of Generalized Linear Models RYAN BAKKER University of Georgia ICPSR Day 2 Bayesian Methods: GLM [1] Likelihood and Maximum Likelihood Principles Likelihood theory is an important part of Bayesian inference: it is how the data enter the model. • The basis is Fisher’s principle: what value of the unknown parameter is “most likely” to have • generated the observed data. Example: flip a coin 10 times, get 5 heads. MLE for p is 0.5. • This is easily the most common and well-understood general estimation process. • Bayesian Methods: GLM [2] Starting details: • – Y is a n k design or observation matrix, θ is a k 1 unknown coefficient vector to be esti- × × mated, we want p(θ Y) (joint sampling distribution or posterior) from p(Y θ) (joint probabil- | | ity function). – Define the likelihood function: n L(θ Y) = p(Y θ) | i| i=1 Y which is no longer on the probability metric. – Our goal is the maximum likelihood value of θ: θˆ : L(θˆ Y) L(θ Y) θ Θ | ≥ | ∀ ∈ where Θ is the class of admissable values for θ. Bayesian Methods: GLM [3] Likelihood and Maximum Likelihood Principles (cont.) Its actually easier to work with the natural log of the likelihood function: • `(θ Y) = log L(θ Y) | | We also find it useful to work with the score function, the first derivative of the log likelihood func- • tion with respect to the parameters of interest: ∂ `˙(θ Y) = `(θ Y) | ∂θ | Setting `˙(θ Y) equal to zero and solving gives the MLE: θˆ, the “most likely” value of θ from the • | parameter space Θ treating the observed data as given. -

Generalized Linear Models
Generalized Linear Models A generalized linear model (GLM) consists of three parts. i) The first part is a random variable giving the conditional distribution of a response Yi given the values of a set of covariates Xij. In the original work on GLM’sby Nelder and Wedderburn (1972) this random variable was a member of an exponential family, but later work has extended beyond this class of random variables. ii) The second part is a linear predictor, i = + 1Xi1 + 2Xi2 + + ··· kXik . iii) The third part is a smooth and invertible link function g(.) which transforms the expected value of the response variable, i = E(Yi) , and is equal to the linear predictor: g(i) = i = + 1Xi1 + 2Xi2 + + kXik. ··· As shown in Tables 15.1 and 15.2, both the general linear model that we have studied extensively and the logistic regression model from Chapter 14 are special cases of this model. One property of members of the exponential family of distributions is that the conditional variance of the response is a function of its mean, (), and possibly a dispersion parameter . The expressions for the variance functions for common members of the exponential family are shown in Table 15.2. Also, for each distribution there is a so-called canonical link function, which simplifies some of the GLM calculations, which is also shown in Table 15.2. Estimation and Testing for GLMs Parameter estimation in GLMs is conducted by the method of maximum likelihood. As with logistic regression models from the last chapter, the generalization of the residual sums of squares from the general linear model is the residual deviance, Dm 2(log Ls log Lm), where Lm is the maximized likelihood for the model of interest, and Ls is the maximized likelihood for a saturated model, which has one parameter per observation and fits the data as well as possible. -

Heteroscedastic Errors
Heteroscedastic Errors ◮ Sometimes plots and/or tests show that the error variances 2 σi = Var(ǫi ) depend on i ◮ Several standard approaches to fixing the problem, depending on the nature of the dependence. ◮ Weighted Least Squares. ◮ Transformation of the response. ◮ Generalized Linear Models. Richard Lockhart STAT 350: Heteroscedastic Errors and GLIM Weighted Least Squares ◮ Suppose variances are known except for a constant factor. 2 2 ◮ That is, σi = σ /wi . ◮ Use weighted least squares. (See Chapter 10 in the text.) ◮ This usually arises realistically in the following situations: ◮ Yi is an average of ni measurements where you know ni . Then wi = ni . 2 ◮ Plots suggest that σi might be proportional to some power of 2 γ γ some covariate: σi = kxi . Then wi = xi− . Richard Lockhart STAT 350: Heteroscedastic Errors and GLIM Variances depending on (mean of) Y ◮ Two standard approaches are available: ◮ Older approach is transformation. ◮ Newer approach is use of generalized linear model; see STAT 402. Richard Lockhart STAT 350: Heteroscedastic Errors and GLIM Transformation ◮ Compute Yi∗ = g(Yi ) for some function g like logarithm or square root. ◮ Then regress Yi∗ on the covariates. ◮ This approach sometimes works for skewed response variables like income; ◮ after transformation we occasionally find the errors are more nearly normal, more homoscedastic and that the model is simpler. ◮ See page 130ff and check under transformations and Box-Cox in the index. Richard Lockhart STAT 350: Heteroscedastic Errors and GLIM Generalized Linear Models ◮ Transformation uses the model T E(g(Yi )) = xi β while generalized linear models use T g(E(Yi )) = xi β ◮ Generally latter approach offers more flexibility. -

Generalized Linear Models
CHAPTER 6 Generalized linear models 6.1 Introduction Generalized linear modeling is a framework for statistical analysis that includes linear and logistic regression as special cases. Linear regression directly predicts continuous data y from a linear predictor Xβ = β0 + X1β1 + + Xkβk.Logistic regression predicts Pr(y =1)forbinarydatafromalinearpredictorwithaninverse-··· logit transformation. A generalized linear model involves: 1. A data vector y =(y1,...,yn) 2. Predictors X and coefficients β,formingalinearpredictorXβ 1 3. A link function g,yieldingavectoroftransformeddataˆy = g− (Xβ)thatare used to model the data 4. A data distribution, p(y yˆ) | 5. Possibly other parameters, such as variances, overdispersions, and cutpoints, involved in the predictors, link function, and data distribution. The options in a generalized linear model are the transformation g and the data distribution p. In linear regression,thetransformationistheidentity(thatis,g(u) u)and • the data distribution is normal, with standard deviation σ estimated from≡ data. 1 1 In logistic regression,thetransformationistheinverse-logit,g− (u)=logit− (u) • (see Figure 5.2a on page 80) and the data distribution is defined by the proba- bility for binary data: Pr(y =1)=y ˆ. This chapter discusses several other classes of generalized linear model, which we list here for convenience: The Poisson model (Section 6.2) is used for count data; that is, where each • data point yi can equal 0, 1, 2, ....Theusualtransformationg used here is the logarithmic, so that g(u)=exp(u)transformsacontinuouslinearpredictorXiβ to a positivey ˆi.ThedatadistributionisPoisson. It is usually a good idea to add a parameter to this model to capture overdis- persion,thatis,variationinthedatabeyondwhatwouldbepredictedfromthe Poisson distribution alone. -

Lecture 18: Regression in Practice
Regression in Practice Regression in Practice ● Regression Errors ● Regression Diagnostics ● Data Transformations Regression Errors Ice Cream Sales vs. Temperature Image source Linear Regression in R > summary(lm(sales ~ temp)) Call: lm(formula = sales ~ temp) Residuals: Min 1Q Median 3Q Max -74.467 -17.359 3.085 23.180 42.040 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) -122.988 54.761 -2.246 0.0513 . temp 28.427 2.816 10.096 3.31e-06 *** --- Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 Residual standard error: 35.07 on 9 degrees of freedom Multiple R-squared: 0.9189, Adjusted R-squared: 0.9098 F-statistic: 101.9 on 1 and 9 DF, p-value: 3.306e-06 Some Goodness-of-fit Statistics ● Residual standard error ● R2 and adjusted R2 ● F statistic Anatomy of Regression Errors Image Source Residual Standard Error ● A residual is a difference between a fitted value and an observed value. ● The total residual error (RSS) is the sum of the squared residuals. ○ Intuitively, RSS is the error that the model does not explain. ● It is a measure of how far the data are from the regression line (i.e., the model), on average, expressed in the units of the dependent variable. ● The standard error of the residuals is roughly the square root of the average residual error (RSS / n). ○ Technically, it’s not √(RSS / n), it’s √(RSS / (n - 2)); it’s adjusted by degrees of freedom. R2: Coefficient of Determination ● R2 = ESS / TSS ● Interpretations: ○ The proportion of the variance in the dependent variable that the model explains. -

Generalized Linear Models Outperform Commonly Used Canonical Analysis in Estimating Spatial Structure of Presence/Absence Data
Generalized Linear Models outperform commonly used canonical analysis in estimating spatial structure of presence/absence data Lélis A. Carlos-Júnior1,2,3, Joel C. Creed4, Rob Marrs2, Rob J. Lewis5, Timothy P. Moulton4, Rafael Feijó-Lima1,6 and Matthew Spencer2 1 Programa de Pós-Graduacão¸ em Ecologia e Evolucão,¸ Universidade do Estado do Rio do Janeiro, Rio de Janeiro, Brazil 2 School of Environmental Sciences, University of Liverpool, Liverpool, United Kingdom 3 Departamento de Biologia, Pontifícia Universidade Católica do Rio de Janeiro, Rio de Janeiro, Brazil 4 Departamento de Ecologia, Universidade do Estado do Rio de Janeiro, Rio de Janeiro, Brazil 5 Department of Forest Genetics and Biodiversity, Norwegian Institute of Bioeconomy Research, Bergen, Nor- way 6 Division of Biological Sciences, University of Montana, Missoula, MT, United States of America ABSTRACT Background. Ecological communities tend to be spatially structured due to environ- mental gradients and/or spatially contagious processes such as growth, dispersion and species interactions. Data transformation followed by usage of algorithms such as Redundancy Analysis (RDA) is a fairly common approach in studies searching for spatial structure in ecological communities, despite recent suggestions advocating the use of Generalized Linear Models (GLMs). Here, we compared the performance of GLMs and RDA in describing spatial structure in ecological community composition data. We simulated realistic presence/absence data typical of many β-diversity studies. For model selection we used standard methods commonly used in most studies involving RDA and GLMs. Submitted 19 November 2018 Methods. We simulated communities with known spatial structure, based on three Accepted 30 July 2020 real spatial community presence/absence datasets (one terrestrial, one marine and Published 3 September 2020 one freshwater). -

Chapter 2 Simple Linear Regression Analysis the Simple
Chapter 2 Simple Linear Regression Analysis The simple linear regression model We consider the modelling between the dependent and one independent variable. When there is only one independent variable in the linear regression model, the model is generally termed as a simple linear regression model. When there are more than one independent variables in the model, then the linear model is termed as the multiple linear regression model. The linear model Consider a simple linear regression model yX01 where y is termed as the dependent or study variable and X is termed as the independent or explanatory variable. The terms 0 and 1 are the parameters of the model. The parameter 0 is termed as an intercept term, and the parameter 1 is termed as the slope parameter. These parameters are usually called as regression coefficients. The unobservable error component accounts for the failure of data to lie on the straight line and represents the difference between the true and observed realization of y . There can be several reasons for such difference, e.g., the effect of all deleted variables in the model, variables may be qualitative, inherent randomness in the observations etc. We assume that is observed as independent and identically distributed random variable with mean zero and constant variance 2 . Later, we will additionally assume that is normally distributed. The independent variables are viewed as controlled by the experimenter, so it is considered as non-stochastic whereas y is viewed as a random variable with Ey()01 X and Var() y 2 . Sometimes X can also be a random variable. -

1 Simple Linear Regression I – Least Squares Estimation
1 Simple Linear Regression I – Least Squares Estimation Textbook Sections: 18.1–18.3 Previously, we have worked with a random variable x that comes from a population that is normally distributed with mean µ and variance σ2. We have seen that we can write x in terms of µ and a random error component ε, that is, x = µ + ε. For the time being, we are going to change our notation for our random variable from x to y. So, we now write y = µ + ε. We will now find it useful to call the random variable y a dependent or response variable. Many times, the response variable of interest may be related to the value(s) of one or more known or controllable independent or predictor variables. Consider the following situations: LR1 A college recruiter would like to be able to predict a potential incoming student’s first–year GPA (y) based on known information concerning high school GPA (x1) and college entrance examination score (x2). She feels that the student’s first–year GPA will be related to the values of these two known variables. LR2 A marketer is interested in the effect of changing shelf height (x1) and shelf width (x2)on the weekly sales (y) of her brand of laundry detergent in a grocery store. LR3 A psychologist is interested in testing whether the amount of time to become proficient in a foreign language (y) is related to the child’s age (x). In each case we have at least one variable that is known (in some cases it is controllable), and a response variable that is a random variable. -

Linear Regression Using Stata (V.6.3)
Linear Regression using Stata (v.6.3) Oscar Torres-Reyna [email protected] December 2007 http://dss.princeton.edu/training/ Regression: a practical approach (overview) We use regression to estimate the unknown effect of changing one variable over another (Stock and Watson, 2003, ch. 4) When running a regression we are making two assumptions, 1) there is a linear relationship between two variables (i.e. X and Y) and 2) this relationship is additive (i.e. Y= x1 + x2 + …+xN). Technically, linear regression estimates how much Y changes when X changes one unit. In Stata use the command regress, type: regress [dependent variable] [independent variable(s)] regress y x In a multivariate setting we type: regress y x1 x2 x3 … Before running a regression it is recommended to have a clear idea of what you are trying to estimate (i.e. which are your outcome and predictor variables). A regression makes sense only if there is a sound theory behind it. 2 PU/DSS/OTR Regression: a practical approach (setting) Example: Are SAT scores higher in states that spend more money on education controlling by other factors?* – Outcome (Y) variable – SAT scores, variable csat in dataset – Predictor (X) variables • Per pupil expenditures primary & secondary (expense) • % HS graduates taking SAT (percent) • Median household income (income) • % adults with HS diploma (high) • % adults with college degree (college) • Region (region) *Source: Data and examples come from the book Statistics with Stata (updated for version 9) by Lawrence C. Hamilton (chapter 6). Click here to download the data or search for it at http://www.duxbury.com/highered/.