Bayesian Methods: Review of Generalized Linear Models
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Analysis of Variance; *********43***1****T******T
DOCUMENT RESUME 11 571 TM 007 817 111, AUTHOR Corder-Bolz, Charles R. TITLE A Monte Carlo Study of Six Models of Change. INSTITUTION Southwest Eduoational Development Lab., Austin, Tex. PUB DiTE (7BA NOTE"". 32p. BDRS-PRICE MF-$0.83 Plug P4istage. HC Not Availablefrom EDRS. DESCRIPTORS *Analysis Of COariance; *Analysis of Variance; Comparative Statistics; Hypothesis Testing;. *Mathematicai'MOdels; Scores; Statistical Analysis; *Tests'of Significance ,IDENTIFIERS *Change Scores; *Monte Carlo Method ABSTRACT A Monte Carlo Study was conducted to evaluate ,six models commonly used to evaluate change. The results.revealed specific problems with each. Analysis of covariance and analysis of variance of residualized gain scores appeared to, substantially and consistently overestimate the change effects. Multiple factor analysis of variance models utilizing pretest and post-test scores yielded invalidly toF ratios. The analysis of variance of difference scores an the multiple 'factor analysis of variance using repeated measures were the only models which adequately controlled for pre-treatment differences; ,however, they appeared to be robust only when the error level is 50% or more. This places serious doubt regarding published findings, and theories based upon change score analyses. When collecting data which. have an error level less than 50% (which is true in most situations)r a change score analysis is entirely inadvisable until an alternative procedure is developed. (Author/CTM) 41t i- **************************************4***********************i,******* 4! Reproductions suppliedby ERRS are theibest that cap be made * .... * '* ,0 . from the original document. *********43***1****t******t*******************************************J . S DEPARTMENT OF HEALTH, EDUCATIONWELFARE NATIONAL INST'ITUTE OF r EOUCATIOp PHISDOCUMENT DUCED EXACTLYHAS /3EENREPRC AS RECEIVED THE PERSONOR ORGANIZATION J RoP AT INC. -

Generalized Linear Models (Glms)
San Jos´eState University Math 261A: Regression Theory & Methods Generalized Linear Models (GLMs) Dr. Guangliang Chen This lecture is based on the following textbook sections: • Chapter 13: 13.1 – 13.3 Outline of this presentation: • What is a GLM? • Logistic regression • Poisson regression Generalized Linear Models (GLMs) What is a GLM? In ordinary linear regression, we assume that the response is a linear function of the regressors plus Gaussian noise: 0 2 y = β0 + β1x1 + ··· + βkxk + ∼ N(x β, σ ) | {z } |{z} linear form x0β N(0,σ2) noise The model can be reformulate in terms of • distribution of the response: y | x ∼ N(µ, σ2), and • dependence of the mean on the predictors: µ = E(y | x) = x0β Dr. Guangliang Chen | Mathematics & Statistics, San Jos´e State University3/24 Generalized Linear Models (GLMs) beta=(1,2) 5 4 3 β0 + β1x b y 2 y 1 0 −1 0.0 0.2 0.4 0.6 0.8 1.0 x x Dr. Guangliang Chen | Mathematics & Statistics, San Jos´e State University4/24 Generalized Linear Models (GLMs) Generalized linear models (GLM) extend linear regression by allowing the response variable to have • a general distribution (with mean µ = E(y | x)) and • a mean that depends on the predictors through a link function g: That is, g(µ) = β0x or equivalently, µ = g−1(β0x) Dr. Guangliang Chen | Mathematics & Statistics, San Jos´e State University5/24 Generalized Linear Models (GLMs) In GLM, the response is typically assumed to have a distribution in the exponential family, which is a large class of probability distributions that have pdfs of the form f(x | θ) = a(x)b(θ) exp(c(θ) · T (x)), including • Normal - ordinary linear regression • Bernoulli - Logistic regression, modeling binary data • Binomial - Multinomial logistic regression, modeling general cate- gorical data • Poisson - Poisson regression, modeling count data • Exponential, Gamma - survival analysis Dr. -

6 Modeling Survival Data with Cox Regression Models
CHAPTER 6 ST 745, Daowen Zhang 6 Modeling Survival Data with Cox Regression Models 6.1 The Proportional Hazards Model A proportional hazards model proposed by D.R. Cox (1972) assumes that T z1β1+···+zpβp z β λ(t|z)=λ0(t)e = λ0(t)e , (6.1) where z is a p × 1 vector of covariates such as treatment indicators, prognositc factors, etc., and β is a p × 1 vector of regression coefficients. Note that there is no intercept β0 in model (6.1). Obviously, λ(t|z =0)=λ0(t). So λ0(t) is often called the baseline hazard function. It can be interpreted as the hazard function for the population of subjects with z =0. The baseline hazard function λ0(t) in model (6.1) can take any shape as a function of t.The T only requirement is that λ0(t) > 0. This is the nonparametric part of the model and z β is the parametric part of the model. So Cox’s proportional hazards model is a semiparametric model. Interpretation of a proportional hazards model 1. It is easy to show that under model (6.1) exp(zT β) S(t|z)=[S0(t)] , where S(t|z) is the survival function of the subpopulation with covariate z and S0(t)isthe survival function of baseline population (z = 0). That is R t − λ0(u)du S0(t)=e 0 . PAGE 120 CHAPTER 6 ST 745, Daowen Zhang 2. For any two sets of covariates z0 and z1, zT β λ(t|z1) λ0(t)e 1 T (z1−z0) β, t ≥ , = zT β =e for all 0 λ(t|z0) λ0(t)e 0 which is a constant over time (so the name of proportional hazards model). -

Bayesian Inference Chapter 4: Regression and Hierarchical Models
Bayesian Inference Chapter 4: Regression and Hierarchical Models Conchi Aus´ınand Mike Wiper Department of Statistics Universidad Carlos III de Madrid Master in Business Administration and Quantitative Methods Master in Mathematical Engineering Conchi Aus´ınand Mike Wiper Regression and hierarchical models Masters Programmes 1 / 35 Objective AFM Smith Dennis Lindley We analyze the Bayesian approach to fitting normal and generalized linear models and introduce the Bayesian hierarchical modeling approach. Also, we study the modeling and forecasting of time series. Conchi Aus´ınand Mike Wiper Regression and hierarchical models Masters Programmes 2 / 35 Contents 1 Normal linear models 1.1. ANOVA model 1.2. Simple linear regression model 2 Generalized linear models 3 Hierarchical models 4 Dynamic models Conchi Aus´ınand Mike Wiper Regression and hierarchical models Masters Programmes 3 / 35 Normal linear models A normal linear model is of the following form: y = Xθ + ; 0 where y = (y1;:::; yn) is the observed data, X is a known n × k matrix, called 0 the design matrix, θ = (θ1; : : : ; θk ) is the parameter set and follows a multivariate normal distribution. Usually, it is assumed that: 1 ∼ N 0 ; I : k φ k A simple example of normal linear model is the simple linear regression model T 1 1 ::: 1 where X = and θ = (α; β)T . x1 x2 ::: xn Conchi Aus´ınand Mike Wiper Regression and hierarchical models Masters Programmes 4 / 35 Normal linear models Consider a normal linear model, y = Xθ + . A conjugate prior distribution is a normal-gamma distribution: -

Fast Computation of the Deviance Information Criterion for Latent Variable Models
Crawford School of Public Policy CAMA Centre for Applied Macroeconomic Analysis Fast Computation of the Deviance Information Criterion for Latent Variable Models CAMA Working Paper 9/2014 January 2014 Joshua C.C. Chan Research School of Economics, ANU and Centre for Applied Macroeconomic Analysis Angelia L. Grant Centre for Applied Macroeconomic Analysis Abstract The deviance information criterion (DIC) has been widely used for Bayesian model comparison. However, recent studies have cautioned against the use of the DIC for comparing latent variable models. In particular, the DIC calculated using the conditional likelihood (obtained by conditioning on the latent variables) is found to be inappropriate, whereas the DIC computed using the integrated likelihood (obtained by integrating out the latent variables) seems to perform well. In view of this, we propose fast algorithms for computing the DIC based on the integrated likelihood for a variety of high- dimensional latent variable models. Through three empirical applications we show that the DICs based on the integrated likelihoods have much smaller numerical standard errors compared to the DICs based on the conditional likelihoods. THE AUSTRALIAN NATIONAL UNIVERSITY Keywords Bayesian model comparison, state space, factor model, vector autoregression, semiparametric JEL Classification C11, C15, C32, C52 Address for correspondence: (E) [email protected] The Centre for Applied Macroeconomic Analysis in the Crawford School of Public Policy has been established to build strong links between professional macroeconomists. It provides a forum for quality macroeconomic research and discussion of policy issues between academia, government and the private sector. The Crawford School of Public Policy is the Australian National University’s public policy school, serving and influencing Australia, Asia and the Pacific through advanced policy research, graduate and executive education, and policy impact. -

A Generalized Linear Model for Binomial Response Data
A Generalized Linear Model for Binomial Response Data Copyright c 2017 Dan Nettleton (Iowa State University) Statistics 510 1 / 46 Now suppose that instead of a Bernoulli response, we have a binomial response for each unit in an experiment or an observational study. As an example, consider the trout data set discussed on page 669 of The Statistical Sleuth, 3rd edition, by Ramsey and Schafer. Five doses of toxic substance were assigned to a total of 20 fish tanks using a completely randomized design with four tanks per dose. Copyright c 2017 Dan Nettleton (Iowa State University) Statistics 510 2 / 46 For each tank, the total number of fish and the number of fish that developed liver tumors were recorded. d=read.delim("http://dnett.github.io/S510/Trout.txt") d dose tumor total 1 0.010 9 87 2 0.010 5 86 3 0.010 2 89 4 0.010 9 85 5 0.025 30 86 6 0.025 41 86 7 0.025 27 86 8 0.025 34 88 9 0.050 54 89 10 0.050 53 86 11 0.050 64 90 12 0.050 55 88 13 0.100 71 88 14 0.100 73 89 15 0.100 65 88 16 0.100 72 90 17 0.250 66 86 18 0.250 75 82 19 0.250 72 81 20 0.250 73 89 Copyright c 2017 Dan Nettleton (Iowa State University) Statistics 510 3 / 46 One way to analyze this dataset would be to convert the binomial counts and totals into Bernoulli responses. -

18.650 (F16) Lecture 10: Generalized Linear Models (Glms)
Statistics for Applications Chapter 10: Generalized Linear Models (GLMs) 1/52 Linear model A linear model assumes Y X (µ(X), σ2I), | ∼ N And ⊤ IE(Y X) = µ(X) = X β, | 2/52 Components of a linear model The two components (that we are going to relax) are 1. Random component: the response variable Y X is continuous | and normally distributed with mean µ = µ(X) = IE(Y X). | 2. Link: between the random and covariates X = (X(1),X(2), ,X(p))⊤: µ(X) = X⊤β. · · · 3/52 Generalization A generalized linear model (GLM) generalizes normal linear regression models in the following directions. 1. Random component: Y some exponential family distribution ∼ 2. Link: between the random and covariates: ⊤ g µ(X) = X β where g called link function� and� µ = IE(Y X). | 4/52 Example 1: Disease Occuring Rate In the early stages of a disease epidemic, the rate at which new cases occur can often increase exponentially through time. Hence, if µi is the expected number of new cases on day ti, a model of the form µi = γ exp(δti) seems appropriate. ◮ Such a model can be turned into GLM form, by using a log link so that log(µi) = log(γ) + δti = β0 + β1ti. ◮ Since this is a count, the Poisson distribution (with expected value µi) is probably a reasonable distribution to try. 5/52 Example 2: Prey Capture Rate(1) The rate of capture of preys, yi, by a hunting animal, tends to increase with increasing density of prey, xi, but to eventually level off, when the predator is catching as much as it can cope with. -

Comparison of Some Chemometric Tools for Metabonomics Biomarker Identification ⁎ Réjane Rousseau A, , Bernadette Govaerts A, Michel Verleysen A,B, Bruno Boulanger C
Available online at www.sciencedirect.com Chemometrics and Intelligent Laboratory Systems 91 (2008) 54–66 www.elsevier.com/locate/chemolab Comparison of some chemometric tools for metabonomics biomarker identification ⁎ Réjane Rousseau a, , Bernadette Govaerts a, Michel Verleysen a,b, Bruno Boulanger c a Université Catholique de Louvain, Institut de Statistique, Voie du Roman Pays 20, B-1348 Louvain-la-Neuve, Belgium b Université Catholique de Louvain, Machine Learning Group - DICE, Belgium c Eli Lilly, European Early Phase Statistics, Belgium Received 29 December 2006; received in revised form 15 June 2007; accepted 22 June 2007 Available online 29 June 2007 Abstract NMR-based metabonomics discovery approaches require statistical methods to extract, from large and complex spectral databases, biomarkers or biologically significant variables that best represent defined biological conditions. This paper explores the respective effectiveness of six multivariate methods: multiple hypotheses testing, supervised extensions of principal (PCA) and independent components analysis (ICA), discriminant partial least squares, linear logistic regression and classification trees. Each method has been adapted in order to provide a biomarker score for each zone of the spectrum. These scores aim at giving to the biologist indications on which metabolites of the analyzed biofluid are potentially affected by a stressor factor of interest (e.g. toxicity of a drug, presence of a given disease or therapeutic effect of a drug). The applications of the six methods to samples of 60 and 200 spectra issued from a semi-artificial database allowed to evaluate their respective properties. In particular, their sensitivities and false discovery rates (FDR) are illustrated through receiver operating characteristics curves (ROC) and the resulting identifications are used to show their specificities and relative advantages.The paper recommends to discard two methods for biomarker identification: the PCA showing a general low efficiency and the CART which is very sensitive to noise. -

Optimal Variance Control of the Score Function Gradient Estimator for Importance Weighted Bounds
Optimal Variance Control of the Score Function Gradient Estimator for Importance Weighted Bounds Valentin Liévin 1 Andrea Dittadi1 Anders Christensen1 Ole Winther1, 2, 3 1 Section for Cognitive Systems, Technical University of Denmark 2 Bioinformatics Centre, Department of Biology, University of Copenhagen 3 Centre for Genomic Medicine, Rigshospitalet, Copenhagen University Hospital {valv,adit}@dtu.dk, [email protected], [email protected] Abstract This paper introduces novel results for the score function gradient estimator of the importance weighted variational bound (IWAE). We prove that in the limit of large K (number of importance samples) one can choose the control variate such that the Signal-to-Noise ratio (SNR) of the estimator grows as pK. This is in contrast to the standard pathwise gradient estimator where the SNR decreases as 1/pK. Based on our theoretical findings we develop a novel control variate that extends on VIMCO. Empirically, for the training of both continuous and discrete generative models, the proposed method yields superior variance reduction, resulting in an SNR for IWAE that increases with K without relying on the reparameterization trick. The novel estimator is competitive with state-of-the-art reparameterization-free gradient estimators such as Reweighted Wake-Sleep (RWS) and the thermodynamic variational objective (TVO) when training generative models. 1 Introduction Gradient-based learning is now widespread in the field of machine learning, in which recent advances have mostly relied on the backpropagation algorithm, the workhorse of modern deep learning. In many instances, for example in the context of unsupervised learning, it is desirable to make models more expressive by introducing stochastic latent variables. -
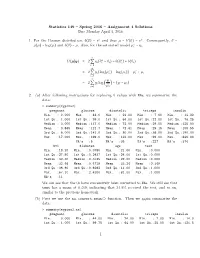
Statistics 149 – Spring 2016 – Assignment 4 Solutions Due Monday April 4, 2016 1. for the Poisson Distribution, B(Θ) =
Statistics 149 { Spring 2016 { Assignment 4 Solutions Due Monday April 4, 2016 1. For the Poisson distribution, b(θ) = eθ and thus µ = b′(θ) = eθ. Consequently, θ = ∗ g(µ) = log(µ) and b(θ) = µ. Also, for the saturated model µi = yi. n ∗ ∗ D(µSy) = 2 Q yi(θi − θi) − b(θi ) + b(θi) i=1 n ∗ ∗ = 2 Q yi(log(µi ) − log(µi)) − µi + µi i=1 n yi = 2 Q yi log − (yi − µi) i=1 µi 2. (a) After following instructions for replacing 0 values with NAs, we summarize the data: > summary(mypima2) pregnant glucose diastolic triceps insulin Min. : 0.000 Min. : 44.0 Min. : 24.00 Min. : 7.00 Min. : 14.00 1st Qu.: 1.000 1st Qu.: 99.0 1st Qu.: 64.00 1st Qu.:22.00 1st Qu.: 76.25 Median : 3.000 Median :117.0 Median : 72.00 Median :29.00 Median :125.00 Mean : 3.845 Mean :121.7 Mean : 72.41 Mean :29.15 Mean :155.55 3rd Qu.: 6.000 3rd Qu.:141.0 3rd Qu.: 80.00 3rd Qu.:36.00 3rd Qu.:190.00 Max. :17.000 Max. :199.0 Max. :122.00 Max. :99.00 Max. :846.00 NA's :5 NA's :35 NA's :227 NA's :374 bmi diabetes age test Min. :18.20 Min. :0.0780 Min. :21.00 Min. :0.000 1st Qu.:27.50 1st Qu.:0.2437 1st Qu.:24.00 1st Qu.:0.000 Median :32.30 Median :0.3725 Median :29.00 Median :0.000 Mean :32.46 Mean :0.4719 Mean :33.24 Mean :0.349 3rd Qu.:36.60 3rd Qu.:0.6262 3rd Qu.:41.00 3rd Qu.:1.000 Max. -

Generalized Linear Models
Generalized Linear Models A generalized linear model (GLM) consists of three parts. i) The first part is a random variable giving the conditional distribution of a response Yi given the values of a set of covariates Xij. In the original work on GLM’sby Nelder and Wedderburn (1972) this random variable was a member of an exponential family, but later work has extended beyond this class of random variables. ii) The second part is a linear predictor, i = + 1Xi1 + 2Xi2 + + ··· kXik . iii) The third part is a smooth and invertible link function g(.) which transforms the expected value of the response variable, i = E(Yi) , and is equal to the linear predictor: g(i) = i = + 1Xi1 + 2Xi2 + + kXik. ··· As shown in Tables 15.1 and 15.2, both the general linear model that we have studied extensively and the logistic regression model from Chapter 14 are special cases of this model. One property of members of the exponential family of distributions is that the conditional variance of the response is a function of its mean, (), and possibly a dispersion parameter . The expressions for the variance functions for common members of the exponential family are shown in Table 15.2. Also, for each distribution there is a so-called canonical link function, which simplifies some of the GLM calculations, which is also shown in Table 15.2. Estimation and Testing for GLMs Parameter estimation in GLMs is conducted by the method of maximum likelihood. As with logistic regression models from the last chapter, the generalization of the residual sums of squares from the general linear model is the residual deviance, Dm 2(log Ls log Lm), where Lm is the maximized likelihood for the model of interest, and Ls is the maximized likelihood for a saturated model, which has one parameter per observation and fits the data as well as possible. -

Simple, Lower-Variance Gradient Estimators for Variational Inference
Sticking the Landing: Simple, Lower-Variance Gradient Estimators for Variational Inference Geoffrey Roeder Yuhuai Wu University of Toronto University of Toronto [email protected] [email protected] David Duvenaud University of Toronto [email protected] Abstract We propose a simple and general variant of the standard reparameterized gradient estimator for the variational evidence lower bound. Specifically, we remove a part of the total derivative with respect to the variational parameters that corresponds to the score function. Removing this term produces an unbiased gradient estimator whose variance approaches zero as the approximate posterior approaches the exact posterior. We analyze the behavior of this gradient estimator theoretically and empirically, and generalize it to more complex variational distributions such as mixtures and importance-weighted posteriors. 1 Introduction Recent advances in variational inference have begun to Optimization using: make approximate inference practical in large-scale latent Path Derivative ) variable models. One of the main recent advances has Total Derivative been the development of variational autoencoders along true φ with the reparameterization trick [Kingma and Welling, k 2013, Rezende et al., 2014]. The reparameterization init trick is applicable to most continuous latent-variable mod- φ els, and usually provides lower-variance gradient esti- ( mates than the more general REINFORCE gradient es- KL timator [Williams, 1992]. Intuitively, the reparameterization trick provides more in- 400 600 800 1000 1200 formative gradients by exposing the dependence of sam- Iterations pled latent variables z on variational parameters φ. In contrast, the REINFORCE gradient estimate only de- Figure 1: Fitting a 100-dimensional varia- pends on the relationship between the density function tional posterior to another Gaussian, using log qφ(z x; φ) and its parameters.