Robustbase: Basic Robust Statistics
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Lecture 12 Robust Estimation
Lecture 12 Robust Estimation Prof. Dr. Svetlozar Rachev Institute for Statistics and Mathematical Economics University of Karlsruhe Financial Econometrics, Summer Semester 2007 Prof. Dr. Svetlozar Rachev Institute for Statistics and MathematicalLecture Economics 12 Robust University Estimation of Karlsruhe Copyright These lecture-notes cannot be copied and/or distributed without permission. The material is based on the text-book: Financial Econometrics: From Basics to Advanced Modeling Techniques (Wiley-Finance, Frank J. Fabozzi Series) by Svetlozar T. Rachev, Stefan Mittnik, Frank Fabozzi, Sergio M. Focardi,Teo Jaˇsic`. Prof. Dr. Svetlozar Rachev Institute for Statistics and MathematicalLecture Economics 12 Robust University Estimation of Karlsruhe Outline I Robust statistics. I Robust estimators of regressions. I Illustration: robustness of the corporate bond yield spread model. Prof. Dr. Svetlozar Rachev Institute for Statistics and MathematicalLecture Economics 12 Robust University Estimation of Karlsruhe Robust Statistics I Robust statistics addresses the problem of making estimates that are insensitive to small changes in the basic assumptions of the statistical models employed. I The concepts and methods of robust statistics originated in the 1950s. However, the concepts of robust statistics had been used much earlier. I Robust statistics: 1. assesses the changes in estimates due to small changes in the basic assumptions; 2. creates new estimates that are insensitive to small changes in some of the assumptions. I Robust statistics is also useful to separate the contribution of the tails from the contribution of the body of the data. Prof. Dr. Svetlozar Rachev Institute for Statistics and MathematicalLecture Economics 12 Robust University Estimation of Karlsruhe Robust Statistics I Peter Huber observed, that robust, distribution-free, and nonparametrical actually are not closely related properties. -

Robust Statistics Part 3: Regression Analysis
Robust Statistics Part 3: Regression analysis Peter Rousseeuw LARS-IASC School, May 2019 Peter Rousseeuw Robust Statistics, Part 3: Regression LARS-IASC School, May 2019 p. 1 Linear regression Linear regression: Outline 1 Classical regression estimators 2 Classical outlier diagnostics 3 Regression M-estimators 4 The LTS estimator 5 Outlier detection 6 Regression S-estimators and MM-estimators 7 Regression with categorical predictors 8 Software Peter Rousseeuw Robust Statistics, Part 3: Regression LARS-IASC School, May 2019 p. 2 Linear regression Classical estimators The linear regression model The linear regression model says: yi = β0 + β1xi1 + ... + βpxip + εi ′ = xiβ + εi 2 ′ ′ with i.i.d. errors εi ∼ N(0,σ ), xi = (1,xi1,...,xip) and β =(β0,β1,...,βp) . ′ Denote the n × (p + 1) matrix containing the predictors xi as X =(x1,..., xn) , ′ ′ the vector of responses y =(y1,...,yn) and the error vector ε =(ε1,...,εn) . Then: y = Xβ + ε Any regression estimate βˆ yields fitted values yˆ = Xβˆ and residuals ri = ri(βˆ)= yi − yˆi . Peter Rousseeuw Robust Statistics, Part 3: Regression LARS-IASC School, May 2019 p. 3 Linear regression Classical estimators The least squares estimator Least squares estimator n ˆ 2 βLS = argmin ri (β) β i=1 X If X has full rank, then the solution is unique and given by ˆ ′ −1 ′ βLS =(X X) X y The usual unbiased estimator of the error variance is n 1 σˆ2 = r2(βˆ ) LS n − p − 1 i LS i=1 X Peter Rousseeuw Robust Statistics, Part 3: Regression LARS-IASC School, May 2019 p. 4 Linear regression Classical estimators Outliers in regression Different types of outliers: vertical outlier good leverage point • • y • • • regular data • ••• • •• ••• • • • • • • • • • bad leverage point • • •• • x Peter Rousseeuw Robust Statistics, Part 3: Regression LARS-IASC School, May 2019 p. -

Fast Computation of the Deviance Information Criterion for Latent Variable Models
Crawford School of Public Policy CAMA Centre for Applied Macroeconomic Analysis Fast Computation of the Deviance Information Criterion for Latent Variable Models CAMA Working Paper 9/2014 January 2014 Joshua C.C. Chan Research School of Economics, ANU and Centre for Applied Macroeconomic Analysis Angelia L. Grant Centre for Applied Macroeconomic Analysis Abstract The deviance information criterion (DIC) has been widely used for Bayesian model comparison. However, recent studies have cautioned against the use of the DIC for comparing latent variable models. In particular, the DIC calculated using the conditional likelihood (obtained by conditioning on the latent variables) is found to be inappropriate, whereas the DIC computed using the integrated likelihood (obtained by integrating out the latent variables) seems to perform well. In view of this, we propose fast algorithms for computing the DIC based on the integrated likelihood for a variety of high- dimensional latent variable models. Through three empirical applications we show that the DICs based on the integrated likelihoods have much smaller numerical standard errors compared to the DICs based on the conditional likelihoods. THE AUSTRALIAN NATIONAL UNIVERSITY Keywords Bayesian model comparison, state space, factor model, vector autoregression, semiparametric JEL Classification C11, C15, C32, C52 Address for correspondence: (E) [email protected] The Centre for Applied Macroeconomic Analysis in the Crawford School of Public Policy has been established to build strong links between professional macroeconomists. It provides a forum for quality macroeconomic research and discussion of policy issues between academia, government and the private sector. The Crawford School of Public Policy is the Australian National University’s public policy school, serving and influencing Australia, Asia and the Pacific through advanced policy research, graduate and executive education, and policy impact. -

A Generalized Linear Model for Binomial Response Data
A Generalized Linear Model for Binomial Response Data Copyright c 2017 Dan Nettleton (Iowa State University) Statistics 510 1 / 46 Now suppose that instead of a Bernoulli response, we have a binomial response for each unit in an experiment or an observational study. As an example, consider the trout data set discussed on page 669 of The Statistical Sleuth, 3rd edition, by Ramsey and Schafer. Five doses of toxic substance were assigned to a total of 20 fish tanks using a completely randomized design with four tanks per dose. Copyright c 2017 Dan Nettleton (Iowa State University) Statistics 510 2 / 46 For each tank, the total number of fish and the number of fish that developed liver tumors were recorded. d=read.delim("http://dnett.github.io/S510/Trout.txt") d dose tumor total 1 0.010 9 87 2 0.010 5 86 3 0.010 2 89 4 0.010 9 85 5 0.025 30 86 6 0.025 41 86 7 0.025 27 86 8 0.025 34 88 9 0.050 54 89 10 0.050 53 86 11 0.050 64 90 12 0.050 55 88 13 0.100 71 88 14 0.100 73 89 15 0.100 65 88 16 0.100 72 90 17 0.250 66 86 18 0.250 75 82 19 0.250 72 81 20 0.250 73 89 Copyright c 2017 Dan Nettleton (Iowa State University) Statistics 510 3 / 46 One way to analyze this dataset would be to convert the binomial counts and totals into Bernoulli responses. -

Comparison of Some Chemometric Tools for Metabonomics Biomarker Identification ⁎ Réjane Rousseau A, , Bernadette Govaerts A, Michel Verleysen A,B, Bruno Boulanger C
Available online at www.sciencedirect.com Chemometrics and Intelligent Laboratory Systems 91 (2008) 54–66 www.elsevier.com/locate/chemolab Comparison of some chemometric tools for metabonomics biomarker identification ⁎ Réjane Rousseau a, , Bernadette Govaerts a, Michel Verleysen a,b, Bruno Boulanger c a Université Catholique de Louvain, Institut de Statistique, Voie du Roman Pays 20, B-1348 Louvain-la-Neuve, Belgium b Université Catholique de Louvain, Machine Learning Group - DICE, Belgium c Eli Lilly, European Early Phase Statistics, Belgium Received 29 December 2006; received in revised form 15 June 2007; accepted 22 June 2007 Available online 29 June 2007 Abstract NMR-based metabonomics discovery approaches require statistical methods to extract, from large and complex spectral databases, biomarkers or biologically significant variables that best represent defined biological conditions. This paper explores the respective effectiveness of six multivariate methods: multiple hypotheses testing, supervised extensions of principal (PCA) and independent components analysis (ICA), discriminant partial least squares, linear logistic regression and classification trees. Each method has been adapted in order to provide a biomarker score for each zone of the spectrum. These scores aim at giving to the biologist indications on which metabolites of the analyzed biofluid are potentially affected by a stressor factor of interest (e.g. toxicity of a drug, presence of a given disease or therapeutic effect of a drug). The applications of the six methods to samples of 60 and 200 spectra issued from a semi-artificial database allowed to evaluate their respective properties. In particular, their sensitivities and false discovery rates (FDR) are illustrated through receiver operating characteristics curves (ROC) and the resulting identifications are used to show their specificities and relative advantages.The paper recommends to discard two methods for biomarker identification: the PCA showing a general low efficiency and the CART which is very sensitive to noise. -
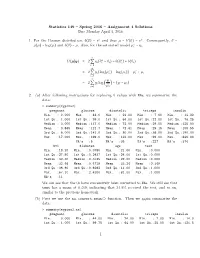
Statistics 149 – Spring 2016 – Assignment 4 Solutions Due Monday April 4, 2016 1. for the Poisson Distribution, B(Θ) =
Statistics 149 { Spring 2016 { Assignment 4 Solutions Due Monday April 4, 2016 1. For the Poisson distribution, b(θ) = eθ and thus µ = b′(θ) = eθ. Consequently, θ = ∗ g(µ) = log(µ) and b(θ) = µ. Also, for the saturated model µi = yi. n ∗ ∗ D(µSy) = 2 Q yi(θi − θi) − b(θi ) + b(θi) i=1 n ∗ ∗ = 2 Q yi(log(µi ) − log(µi)) − µi + µi i=1 n yi = 2 Q yi log − (yi − µi) i=1 µi 2. (a) After following instructions for replacing 0 values with NAs, we summarize the data: > summary(mypima2) pregnant glucose diastolic triceps insulin Min. : 0.000 Min. : 44.0 Min. : 24.00 Min. : 7.00 Min. : 14.00 1st Qu.: 1.000 1st Qu.: 99.0 1st Qu.: 64.00 1st Qu.:22.00 1st Qu.: 76.25 Median : 3.000 Median :117.0 Median : 72.00 Median :29.00 Median :125.00 Mean : 3.845 Mean :121.7 Mean : 72.41 Mean :29.15 Mean :155.55 3rd Qu.: 6.000 3rd Qu.:141.0 3rd Qu.: 80.00 3rd Qu.:36.00 3rd Qu.:190.00 Max. :17.000 Max. :199.0 Max. :122.00 Max. :99.00 Max. :846.00 NA's :5 NA's :35 NA's :227 NA's :374 bmi diabetes age test Min. :18.20 Min. :0.0780 Min. :21.00 Min. :0.000 1st Qu.:27.50 1st Qu.:0.2437 1st Qu.:24.00 1st Qu.:0.000 Median :32.30 Median :0.3725 Median :29.00 Median :0.000 Mean :32.46 Mean :0.4719 Mean :33.24 Mean :0.349 3rd Qu.:36.60 3rd Qu.:0.6262 3rd Qu.:41.00 3rd Qu.:1.000 Max. -

Bayesian Methods: Review of Generalized Linear Models
Bayesian Methods: Review of Generalized Linear Models RYAN BAKKER University of Georgia ICPSR Day 2 Bayesian Methods: GLM [1] Likelihood and Maximum Likelihood Principles Likelihood theory is an important part of Bayesian inference: it is how the data enter the model. • The basis is Fisher’s principle: what value of the unknown parameter is “most likely” to have • generated the observed data. Example: flip a coin 10 times, get 5 heads. MLE for p is 0.5. • This is easily the most common and well-understood general estimation process. • Bayesian Methods: GLM [2] Starting details: • – Y is a n k design or observation matrix, θ is a k 1 unknown coefficient vector to be esti- × × mated, we want p(θ Y) (joint sampling distribution or posterior) from p(Y θ) (joint probabil- | | ity function). – Define the likelihood function: n L(θ Y) = p(Y θ) | i| i=1 Y which is no longer on the probability metric. – Our goal is the maximum likelihood value of θ: θˆ : L(θˆ Y) L(θ Y) θ Θ | ≥ | ∀ ∈ where Θ is the class of admissable values for θ. Bayesian Methods: GLM [3] Likelihood and Maximum Likelihood Principles (cont.) Its actually easier to work with the natural log of the likelihood function: • `(θ Y) = log L(θ Y) | | We also find it useful to work with the score function, the first derivative of the log likelihood func- • tion with respect to the parameters of interest: ∂ `˙(θ Y) = `(θ Y) | ∂θ | Setting `˙(θ Y) equal to zero and solving gives the MLE: θˆ, the “most likely” value of θ from the • | parameter space Θ treating the observed data as given. -

Robustness of Parametric and Nonparametric Tests Under Non-Normality for Two Independent Sample
International Journal of Management and Applied Science, ISSN: 2394-7926 Volume-4, Issue-4, Apr.-2018 http://iraj.in ROBUSTNESS OF PARAMETRIC AND NONPARAMETRIC TESTS UNDER NON-NORMALITY FOR TWO INDEPENDENT SAMPLE 1USMAN, M., 2IBRAHIM, N 1Dept of Statistics, Fed. Polytechnic Bali, Taraba State, Dept of Maths and Statistcs, Fed. Polytechnic MubiNigeria E-mail: [email protected] Abstract - Robust statistical methods have been developed for many common problems, such as estimating location, scale and regression parameters. One motivation is to provide methods with good performance when there are small departures from parametric distributions. This study was aimed to investigates the performance oft-test, Mann Whitney U test and Kolmogorov Smirnov test procedures on independent samples from unrelated population, under situations where the basic assumptions of parametric are not met for different sample size. Testing hypothesis on equality of means require assumptions to be made about the format of the data to be employed. Sometimes the test may depend on the assumption that a sample comes from a distribution in a particular family; if there is a doubt, then a non-parametric tests like Mann Whitney U test orKolmogorov Smirnov test is employed. Random samples were simulated from Normal, Uniform, Exponential, Beta and Gamma distributions. The three tests procedures were applied on the simulated data sets at various sample sizes (small and moderate) and their Type I error and power of the test were studied in both situations under study. Keywords - Non-normal,Independent Sample, T-test,Mann Whitney U test and Kolmogorov Smirnov test. I. INTRODUCTION assumptions about your data, but it may also require the data to be an independent random sample4. -

Robust Methods in Biostatistics
Robust Methods in Biostatistics Stephane Heritier The George Institute for International Health, University of Sydney, Australia Eva Cantoni Department of Econometrics, University of Geneva, Switzerland Samuel Copt Merck Serono International, Geneva, Switzerland Maria-Pia Victoria-Feser HEC Section, University of Geneva, Switzerland A John Wiley and Sons, Ltd, Publication Robust Methods in Biostatistics WILEY SERIES IN PROBABILITY AND STATISTICS Established by WALTER A. SHEWHART and SAMUEL S. WILKS Editors David J. Balding, Noel A. C. Cressie, Garrett M. Fitzmaurice, Iain M. Johnstone, Geert Molenberghs, David W. Scott, Adrian F. M. Smith, Ruey S. Tsay, Sanford Weisberg, Harvey Goldstein. Editors Emeriti Vic Barnett, J. Stuart Hunter, Jozef L. Teugels A complete list of the titles in this series appears at the end of this volume. Robust Methods in Biostatistics Stephane Heritier The George Institute for International Health, University of Sydney, Australia Eva Cantoni Department of Econometrics, University of Geneva, Switzerland Samuel Copt Merck Serono International, Geneva, Switzerland Maria-Pia Victoria-Feser HEC Section, University of Geneva, Switzerland A John Wiley and Sons, Ltd, Publication This edition first published 2009 c 2009 John Wiley & Sons Ltd Registered office John Wiley & Sons Ltd, The Atrium, Southern Gate, Chichester, West Sussex, PO19 8SQ, United Kingdom For details of our global editorial offices, for customer services and for information about how to apply for permission to reuse the copyright material in this book please see our website at www.wiley.com. The right of the author to be identified as the author of this work has been asserted in accordance with the Copyright, Designs and Patents Act 1988. -

A Guide to Robust Statistical Methods in Neuroscience
A GUIDE TO ROBUST STATISTICAL METHODS IN NEUROSCIENCE Authors: Rand R. Wilcox1∗, Guillaume A. Rousselet2 1. Dept. of Psychology, University of Southern California, Los Angeles, CA 90089-1061, USA 2. Institute of Neuroscience and Psychology, College of Medical, Veterinary and Life Sciences, University of Glasgow, 58 Hillhead Street, G12 8QB, Glasgow, UK ∗ Corresponding author: [email protected] ABSTRACT There is a vast array of new and improved methods for comparing groups and studying associations that offer the potential for substantially increasing power, providing improved control over the probability of a Type I error, and yielding a deeper and more nuanced understanding of data. These new techniques effectively deal with four insights into when and why conventional methods can be unsatisfactory. But for the non-statistician, the vast array of new and improved techniques for comparing groups and studying associations can seem daunting, simply because there are so many new methods that are now available. The paper briefly reviews when and why conventional methods can have relatively low power and yield misleading results. The main goal is to suggest some general guidelines regarding when, how and why certain modern techniques might be used. Keywords: Non-normality, heteroscedasticity, skewed distributions, outliers, curvature. 1 1 Introduction The typical introductory statistics course covers classic methods for comparing groups (e.g., Student's t-test, the ANOVA F test and the Wilcoxon{Mann{Whitney test) and studying associations (e.g., Pearson's correlation and least squares regression). The two-sample Stu- dent's t-test and the ANOVA F test assume that sampling is from normal distributions and that the population variances are identical, which is generally known as the homoscedastic- ity assumption. -

What I Should Have Described in Class Today
Statistics 849 Clarification 2010-11-03 (p. 1) What I should have described in class today Sorry for botching the description of the deviance calculation in a generalized linear model and how it relates to the function dev.resids in a glm family object in R. Once again Chen Zuo pointed out the part that I had missed. We need to use positive contributions to the deviance for each observation if we are later to take the square roots of these quantities. To ensure that these are all positive we establish a baseline level for the deviance, which is the lowest possible value of the deviance, corresponding to a saturated model in which each observation has one parameter associated with it. Another value quoted in the summary output is the null deviance, which is the deviance for the simplest possible model (the \null model") corresponding to a formula like > data(Contraception, package = "mlmRev") > summary(cm0 <- glm(use ~ 1, binomial, Contraception)) Call: glm(formula = use ~ 1, family = binomial, data = Contraception) Deviance Residuals: Min 1Q Median 3Q Max -0.9983 -0.9983 -0.9983 1.3677 1.3677 Coefficients: Estimate Std. Error z value Pr(>|z|) (Intercept) -0.43702 0.04657 -9.385 <2e-16 (Dispersion parameter for binomial family taken to be 1) Null deviance: 2590.9 on 1933 degrees of freedom Residual deviance: 2590.9 on 1933 degrees of freedom AIC: 2592.9 Number of Fisher Scoring iterations: 4 Notice that the coefficient estimate, -0.437022, is the logit transformation of the proportion of positive responses > str(y <- as.integer(Contraception[["use"]]) - 1L) int [1:1934] 0 0 0 0 0 0 0 0 0 0 .. -

Robust Statistical Methods in R Using the WRS2 Package
JSS Journal of Statistical Software MMMMMM YYYY, Volume VV, Issue II. doi: 10.18637/jss.v000.i00 Robust Statistical Methods in R Using the WRS2 Package Patrick Mair Rand Wilcox Harvard University University of Southern California Abstract In this manuscript we present various robust statistical methods popular in the social sciences, and show how to apply them in R using the WRS2 package available on CRAN. We elaborate on robust location measures, and present robust t-test and ANOVA ver- sions for independent and dependent samples, including quantile ANOVA. Furthermore, we present on running interval smoothers as used in robust ANCOVA, strategies for com- paring discrete distributions, robust correlation measures and tests, and robust mediator models. Keywords: robust statistics, robust location measures, robust ANOVA, robust ANCOVA, robust mediation, robust correlation. 1. Introduction Data are rarely normal. Yet many classical approaches in inferential statistics assume nor- mally distributed data, especially when it comes to small samples. For large samples the central limit theorem basically tells us that we do not have to worry too much. Unfortu- nately, things are much more complex than that, especially in the case of prominent, \dan- gerous" normality deviations such as skewed distributions, data with outliers, or heavy-tailed distributions. Before elaborating on consequences of these violations within the context of statistical testing and estimation, let us look at the impact of normality deviations from a purely descriptive angle. It is trivial that the mean can be heavily affected by outliers or highly skewed distribu- tional shapes. Computing the mean on such data would not give us the \typical" participant; it is just not a good location measure to characterize the sample.