Myxoma Virus-Encoded Host Range Protein M029: a Multifunctional Antagonist Targeting Multiple Host Antiviral and Innate Immune Pathways
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

How Much Do You Know About Fleas, Ticks, Mites and Other Biters? by Vet Glen Cousquer C Norris
Artful arthropods How much do you know about fleas, ticks, mites and other biters? By vet Glen Cousquer C Norris robably the most historically significant and well known arthropod-borne disease was the “Black Death”. This scourge of the Middle Ages, also known as the Bubonic Plague, killed Pbetween 30 and 60 per cent of Europe’s human population during the 14th Century. This bacterial disease was carried by black rats (Rattus rattus) and transmitted from the rat to humans by the Oriental rat flea (Xenopsylla cheopis). The flea was able to pick up the disease when feeding on rats and then transfer Cheyletiella mites result in the appearance the disease when feeding again on humans. At the time, the of scurf and bald patches in the fur various contributing causes of this devastating disease remained G Cousquer obscure. Our ancestors were largely ignorant of the role played transmit diseases to rabbits. Many of the most important rabbit by the rat, the flea, poor hygiene and inadequate sanitation and diseases are transmitted in this way and we need a detailed this severely hampered their attempts to deal with outbreaks. understanding of the process involved if we are to protect our Today, it is this understanding of how disease can be carried rabbits’ health and welfare. by reservoirs and then transmitted by vectors that allows us to mount more effective responses to such disease threats. So, what are the common arthropod parasites found feeding on rabbits? This feature will look at how certain arthropods play key roles in the transmission of diseases to rabbits. -

For the Riparian Brush Rabbit (Sylvilagus Bachmani Riparius)
CONTROLLED PROPAGATION AND REINTRODUCTION PLAN FOR THE RIPARIAN BRUSH RABBIT (SYLVILAGUS BACHMANI RIPARIUS) Ó B. Moose Peterson, Wildlife Research Photography by DANIEL F. WILLIAMS, PATRICK A. KELLY, AND LAURISSA P. HAMILTON ENDANGERED SPECIES RECOVERY PROGRAM CALIFORNIA STATE UNIVERSITY, STANISLAUS TURLOCK, CA 95382 6 JULY 2002 EXECUTIVE SUMMARY We present a plan for controlled propagation and reintroduction of riparian brush rab- bits (Sylvilagus bachmani riparius), a necessary set of tasks for its recovery, as called for in the Recovery Plan for Upland Species of the San Joaquin Valley, California1. This controlled propagation and reintroduction plan follows the criteria and recommendations of the U.S. Fish and Wildlife Service’s Policy Regarding Controlled Propagation of Spe- cies Listed under the Endangered Species Act2. It is organized along the lines recom- mended in the draft version of that policy and meets the criteria of the final policy. It should be viewed in an adaptive management context in that as events unfold, unexpected changes and new information will require modifications. Modifications will be appended to this document. Controlled Propagation is necessary for the riparian brush rabbit 1) to provide a source of individuals for reintroduction to restored habitat for establishing new, self- sustaining populations, 2) to augment existing populations if needed, 3) and to ensure the prevention of extinction of the species in the wild. We propose to establish three breed- ing colonies in separate enclosures of 1.2-1.4 acres each. Predator-resistant enclosures will be erected around existing, but unoccupied natural habitat for brush rabbits. Enclo- sures will be located on State land surrounded by irrigated agriculture that provides no habitat for brush rabbits. -

TICKS in RELATION to HUMAN DISEASES CAUSED by <I
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln U.S. Navy Research U.S. Department of Defense 1967 TICKS IN RELATION TO HUMAN DISEASES CAUSED BY RICKETTSIA SPECIES Harry Hoogstraal Follow this and additional works at: https://digitalcommons.unl.edu/usnavyresearch This Article is brought to you for free and open access by the U.S. Department of Defense at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in U.S. Navy Research by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. TICKS IN RELATION TO HUMAN DISEASES CAUSED BY RICKETTSIA SPECIES1,2 By HARRY HOOGSTRAAL Department oj Medical Zoology, United States Naval Medical Research Unit Number Three, Cairo, Egypt, U.A.R. Rickettsiae (185) are obligate intracellular parasites that multiply by binary fission in the cells of both vertebrate and invertebrate hosts. They are pleomorphic coccobacillary bodies with complex cell walls containing muramic acid, and internal structures composed of ribonucleic and deoxyri bonucleic acids. Rickettsiae show independent metabolic activity with amino acids and intermediate carbohydrates as substrates, and are very susceptible to tetracyclines as well as to other antibiotics. They may be considered as fastidious bacteria whose major unique character is their obligate intracellu lar life, although there is at least one exception to this. In appearance, they range from coccoid forms 0.3 J.I. in diameter to long chains of bacillary forms. They are thus intermediate in size between most bacteria and filterable viruses, and form the family Rickettsiaceae Pinkerton. They stain poorly by Gram's method but well by the procedures of Macchiavello, Gimenez, and Giemsa. -

WHAT IS MYXOMATOSIS? After Its Introduction to the UK in the Early 1950’S Myxomatosis Was So Virulent That an Estimated 95% of Wild and Pet Rabbits Had Died by 1955
WHAT IS MYXOMATOSIS? After its introduction to the UK in the early 1950’s myxomatosis was so virulent that an estimated 95% of wild and pet rabbits had died by 1955. In the year 2000 we again experienced one of the worst outbreaks of Myxomatosis ever recorded, and this disease is still affecting rabbits, particularly during the late summer/ autumn season. Below: edited extracts from an article written by Dr Linda Dykes, published in Fur & Feather’s November 2000 issue. yxomatosis is a rabbit will be a pitiful sight. Severe viral disease which conjunctivitis causes blindness and decimated the wild is accompanied by swelling of the Mrabbit population when it head and genital region plus lumps arrived in Britain nearly seventy on the body. years ago. The number and The rabbit can take a fortnight to severity of outbreaks varies over die and treatment of the “classic” time as the myxomatosis virus form of myxomatosis is usually is notorious for its ability to futile. mutate from year to year, and the background immunity in There are also two atypical forms the wild rabbit population also of myxomatosis: one causes varies. pneumonia and a snuffles-like illness; the other (“nodular Be especially careful if you have a being a bit “off colour” with a few Rabbits at myxomatosis”) mainly affects skin Greatest Risk dog or cat that hunts wild rabbits, skin lesions. and carries a better prognosis. Domestic rabbits do not have as they could bring rabbit fleas Treatment is usually successful any genetically based immunity If a vaccinated rabbit does develop home or into the rabbitry on their in the vaccinated rabbit with a against myxomatosis. -

Myxomatosis: the Transmission of a Highly Virulent Strain of Myxoma Virus by the European Rabbit Flea Spilopsyllus Cuniculi (Dale) in the Mallee Region of Victoria
J. %, Camb. (1977), 79, 405 405 Printed in Great Britain Myxomatosis: the transmission of a highly virulent strain of myxoma virus by the European rabbit flea Spilopsyllus cuniculi (Dale) in the Mallee region of Victoria BY ROSAMOND C. H. SHEPHERD AND J. W. EDMONDS Keith Turnbull Research Institute, Vermin and Noxious Weeds Destruction Board, Department of Crown Lands and Survey, Franhston, Victoria 3199, Australia (Received 29 March 1977) SUMMARY The European rabbit flea Spilopsyllus cuniculi (Dale) was introduced into Australia to act as a vector of myxoma virus. It was first released in the semi-arid Mallee region of Victoria in 1970 where epizootics caused by field strains of myxoma virus occur each summer. Introductions of the readily identified Lausanne strain were made annually following the release of the flea. The introductions were successful and the strain persisted for up to 16 weeks despite competition from field strains. The Lausanne strain is more readily spread by fleas than the Glenfield strain which has been widely used in rabbit control. The ability of the Lausanne strain to persist and its effective transmission compared with the Glenfield strain may be due in part to the more florid symptoms of the disease. INTRODUCTION When myxoma virus was first introduced into Australia in 1950 little was known about the transmission of the virus from rabbit to rabbit although Bull & Mules (1944) had stressed the necessity for the presence of insect vectors. The role of certain species of mosquito vectors became apparent when the first epizootics occurred and it was established that myxomatosis would develop in association with local and seasonal vector activity (Fenner & Ratcliffe, 1965). -

Classification of the Hares and Their Allies
1 CLASSIFICATION OF THE HARES AND THEIR ALLIES By MARCUS WARD LYON, Jr. CONTENTS I. Introduction 322 II. List of names that have been applied to the existing Hares, Rab- bits, and Pikas 325 III. List of the existing species of the DupHcidentata, arranged by genera and subgenera 334 IV. General consideration of the skeleton and teeth of the DupH- cidentata 337 V. Table showing principal osteological differences between the fam- ilies Ochotonida: and Leporidje 384 VI. Keys to Families, genera, and subgenera of DupHcidentata Based primarily on dental characters 386 Based primarily on cranial characters 387 Based primarily on skeletal characters other than those of the skull 389 VII. Detailed account of the genera and subgenera of the existing Hares, Rabbits, and Pikas 389 Family Leporidae Genus Lepits 389 Subgenus Lepus 394 Subgenus Poccilolagus 395 Subgenus Macrofolagiis 395 Genus Sylvilagus 396 Subgenus Sylvilagus 401 Subgenus Microlagus 402 Genus Oryctolagus 402 Genus Linuiolagiis 406 Genus Bracliylagus 4" Genus Pronolagus 416 Genus Romerolagus 420 Genus Nesolagiis 425 Genus Caprolagiis 426 Genus Pentalagus 428 Family Ochotonidae 43 Genus Ochotona 43' Subgenus Ochotona 43^ Subgenus Conothoa 43^ Subgenus Pika 43^ VIII. Geographical distribution 439 IX. Bibliography 44° X. Explanation of plates 443 321 : 32 2 SMITHSONIAN MISCELLANEOUS COLLECTIONS [vOL. 45 I. INTRODUCTION The object of this paper is to give an account of the principal osteological features of the hares, rabbits, and pikas or duphcidentate rodents, the DnpHcidentata, and to determine their family, generic, and subgeneric relationships. The subject is treated in two ways. First, there is a discussion of each part of the skeleton and of the variations that are found in that part throughout the various groups of the existing Dupliciden- tata. -

Technical Glossary
WBVGL 6/28/03 12:00 AM Page 409 Technical Glossary abortive infection: Infection of a cell where there is no net increase in the production of infectious virus. abortive transformation: See transitory (transient or abortive) transformation. acid blob activator: A regulatory protein that acts in trans to alter gene expression and whose activity depends on a region of an amino acid sequence containing acidic or phosphorylated residues. acquired immune deficiency syndrome (AIDS): A disease characterized by loss of cell-mediated and humoral immunity as the result of infection with human immunodeficiency virus (HIV). acute infection: An infection marked by a sudden onset of detectable symptoms usually followed by complete or apparent recovery. adaptive immunity (acquired immunity): See immunity. adjuvant: Something added to a drug to increase the effectiveness of that drug. With respect to the immune system, an adjuvant increases the response of the system to a particular antigen. agnogene: A region of a genome that contains an open reading frame of unknown function; origi- nally used to describe a 67- to 71-amino acid product from the late region of SV40. AIDS: See acquired immune deficiency syndrome. aliquot: One of a number of replicate samples of known size. a-TIF: The alpha trans-inducing factor protein of HSV; a structural (virion) protein that functions as an acid blob transcriptional activator. Its specificity requires interaction with certain host cel- lular proteins (such as Oct1) that bind to immediate-early promoter enhancers. ambisense genome: An RNA genome that contains sequence information in both the positive and negative senses. The S genomic segment of the Arenaviridae and of certain genera of the Bunyaviridae have this characteristic. -

Cottontail Rabbits
Cottontail Rabbits Biology of Cottontail Rabbits (Sylvilagus spp.) as Prey of Golden Eagles (Aquila chrysaetos) in the Western United States Photo Credit, Sky deLight Credit,Photo Sky Cottontail Rabbits Biology of Cottontail Rabbits (Sylvilagus spp.) as Prey of Golden Eagles (Aquila chrysaetos) in the Western United States U.S. Fish and Wildlife Service Regions 1, 2, 6, and 8 Western Golden Eagle Team Front Matter Date: November 13, 2017 Disclaimer The reports in this series have been prepared by the U.S. Fish and Wildlife Service (Service) Western Golden Eagle Team (WGET) for the purpose of proactively addressing energy-related conservation needs of golden eagles in Regions 1, 2, 6, and 8. The team was composed of Service personnel, sometimes assisted by contractors or outside cooperators. The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the U.S. Fish and Wildlife Service. Suggested Citation Hansen, D.L., G. Bedrosian, and G. Beatty. 2017. Biology of cottontail rabbits (Sylvilagus spp.) as prey of golden eagles (Aquila chrysaetos) in the western United States. Unpublished report prepared by the Western Golden Eagle Team, U.S. Fish and Wildlife Service. Available online at: https://ecos.fws.gov/ServCat/Reference/Profile/87137 Acknowledgments This report was authored by Dan L. Hansen, Geoffrey Bedrosian, and Greg Beatty. The authors are grateful to the following reviewers (in alphabetical order): Katie Powell, Charles R. Preston, and Hillary White. Cottontails—i Summary Cottontail rabbits (Sylvilagus spp.; hereafter, cottontails) are among the most frequently identified prey in the diets of breeding golden eagles (Aquila chrysaetos) in the western United States (U.S.). -

Specimen Type, Collection Methods, and Diagnostic Assays Available For
Specimen type, collection methods, and diagnostic assays available for the detection of poxviruses from human specimens by the Poxvirus and Rabies Branch, Centers for Disease Control and Prevention1. Specimen Orthopoxvirus Parapoxvirus Yatapoxvirus Molluscipoxvirus Specimen type collection method PCR6 Culture EM8 IHC9,10 Serology11 PCR12 EM8 IHC9,10 PCR13 EM8 PCR EM8 Lesion material Fresh or frozen Swab 5 Lesion material [dry or in media ] [vesicle / pustule Formalin fixed skin, scab / crust, etc.] Paraffin block Fixed slide(s) Container Lesion fluid Swab [vesicle / pustule [dry or in media5] fluid, etc.] Touch prep slide Blood EDTA2 EDTA tube 7 Spun or aliquoted Serum before shipment Spun or aliquoted Plasma before shipment CSF3,4 Sterile 1. The detection of poxviruses by electron microscopy (EM) and immunohistochemical staining (IHC) is performed by the Infectious Disease Pathology Branch of the CDC. 2. EDTA — Ethylenediaminetetraacetic acid. 3. CSF — Cerebrospinal fluid. 4. In order to accurately interpret test results generated from CSF specimens, paired serum must also be submitted. 5. If media is used to store and transport specimens a minimal amount should be used to ensure as little dilution of DNA as possible. 6. Orthopoxvirus generic real-time polymerase chain reaction (PCR) assays will amplify DNA from numerous species of virus within the Orthopoxvirus genus. Species-specific real-time PCR assays are available for selective detection of DNA from variola virus, vaccinia virus, monkeypox virus, and cowpox virus. 7. Blood is not ideal for the detection of orthopoxviruses by PCR as the period of viremia has often passed before sampling occurs. 8. EM can reveal the presence of a poxvirus in clinical specimens or from virus culture, but this technique cannot differentiate between virus species within the same genus. -
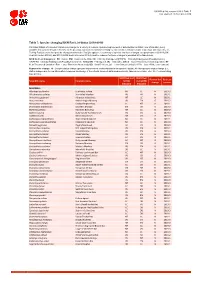
Table 7: Species Changing IUCN Red List Status (2018-2019)
IUCN Red List version 2019-3: Table 7 Last Updated: 10 December 2019 Table 7: Species changing IUCN Red List Status (2018-2019) Published listings of a species' status may change for a variety of reasons (genuine improvement or deterioration in status; new information being available that was not known at the time of the previous assessment; taxonomic changes; corrections to mistakes made in previous assessments, etc. To help Red List users interpret the changes between the Red List updates, a summary of species that have changed category between 2018 (IUCN Red List version 2018-2) and 2019 (IUCN Red List version 2019-3) and the reasons for these changes is provided in the table below. IUCN Red List Categories: EX - Extinct, EW - Extinct in the Wild, CR - Critically Endangered [CR(PE) - Critically Endangered (Possibly Extinct), CR(PEW) - Critically Endangered (Possibly Extinct in the Wild)], EN - Endangered, VU - Vulnerable, LR/cd - Lower Risk/conservation dependent, NT - Near Threatened (includes LR/nt - Lower Risk/near threatened), DD - Data Deficient, LC - Least Concern (includes LR/lc - Lower Risk, least concern). Reasons for change: G - Genuine status change (genuine improvement or deterioration in the species' status); N - Non-genuine status change (i.e., status changes due to new information, improved knowledge of the criteria, incorrect data used previously, taxonomic revision, etc.); E - Previous listing was an Error. IUCN Red List IUCN Red Reason for Red List Scientific name Common name (2018) List (2019) change version Category -

Nscs Are Permissive to Oncolytic Myxoma Virus and Provide a Delivery Method for Targeted Ovarian Cancer Therapy
www.oncotarget.com Oncotarget, 2020, Vol. 11, (No. 51), pp: 4693-4698 Research Paper NSCs are permissive to oncolytic Myxoma virus and provide a delivery method for targeted ovarian cancer therapy Yvonne Cornejo1,2, Min Li3, Thanh H. Dellinger4, Rachael Mooney1, Masmudur M. Rahman5, Grant McFadden5, Karen S. Aboody1,6 and Mohamed Hammad1 1Department of Stem Cell & Developmental Biology, City of Hope, Duarte, CA 91010, USA 2Irell & Manella Graduate School for Biological Sciences, Beckman Research Institute, City of Hope, Duarte, CA 91010, USA 3Department of Information Sciences, Division of Biostatistics at the Beckman Research Institute, City of Hope, Duarte, CA 91010, USA 4Division of Gynecologic Surgery, Department of Surgery, City of Hope, CA 91010, USA 5Biodesign Institute, Arizona State University, Tempe, AZ 85281, USA 6Division of Neurosurgery, City of Hope, Duarte, CA 91010, USA Correspondence to: Mohamed Hammad, email: [email protected] Keywords: oncolytic virotherapy; myxoma; NSCs; ovarian cancer Received: October 09, 2020 Accepted: December 03, 2020 Published: December 22, 2020 Copyright: © 2020 Cornejo et al. This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT Despite the development of many anticancer agents over the past 20 years, ovarian cancer remains the most lethal gynecologic malignancy. Due to a lack of effective screening, the majority of patients with ovarian cancer are diagnosed at an advanced stage, and only ~20% of patients are cured. Thus, in addition to improved screening methods, there is an urgent need for novel anticancer agents that are effective against late-stage, metastatic disease. -

Eastern Cottontail
EASTERN COTTONTAIL This publication is available in alternativeRABBIT media on request. Penn State is an equal opportunity, affirmative action employer, and is committed to providing employment opportunities to all qualified applicants without regard to race, color, religion, age, sex, sexual orientation, gender identity, national origin, disability, or protected veteran status. © The Pennsylvania State University 2017 U.Ed. AGR 17-73 Corner of Park Ave. and Bigler Road • University Park, PA 16802 arboretum.psu.edu facebook.com/pennstatearboretum COTTONTAIL RABBIT DESCRIPTION Named for its characteristic “cotton-ball” tail, the Eastern cottontail is the most widespread species of rabbit in North America. Although most active on rainy or foggy nights, this animal’s brown fur provides excellent camou- NAME: Sylvilagus floridanus flage during the day. Because Eastern cottontails do not hibernate, they can be found in Pennsylvania year-round CONSERVATION STATUS: in open, grassy areas with shrubby cover. The long ears of extinct near least rabbits can move independently, enabling them to hear extinct in wild threatened threatened concern in two directions at once, as well as providing a cooling mechanism through an extensive network of blood vessels. EX EW CR EN VU NT LC DIET SIZE: 1.3–1.5 feet A common visitor to gardens, the Eastern cottontail rabbit enjoys eating grasses, herbs, flowers, fruit, and WEIGHT: 1.75–3.5 pounds vegetables. In the winter, this animal dines on twigs, bark, and plant buds. GROUP TERM: colony; nest THREATS NUMBER OF YOUNG: 3–8 Eastern cottontail rabbits are preyed upon by coyotes, foxes, bobcats, hawks, and even snakes. They may also HABITAT: open fields, meadows be hunted by humans for their meat and fur.