Phylogenetic Analysis of Rhogap Domain-Containing Proteins
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Expression and Localization of Grass Carp Pkc-Θ (Protein Kinase C Theta) Gene After Its Activation T
Fish and Shellfish Immunology 87 (2019) 788–795 Contents lists available at ScienceDirect Fish and Shellfish Immunology journal homepage: www.elsevier.com/locate/fsi Full length article Expression and localization of grass carp pkc-θ (protein kinase C theta) gene after its activation T ∗∗ Rumana Mehjabina,b,1, Liangming Chena,b,1, Rong Huanga, , Denghui Zhua,b, Cheng Yanga,b, ∗ Yongming Lia, Lanjie Liaoa, Libo Hea, Zuoyan Zhua, Yaping Wanga, a State Key Laboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, 430072, China b University of Chinese Academy of Sciences, Beijing, 100049, China ARTICLE INFO ABSTRACT Keywords: Haemorrhagic disease caused by grass carp reovirus (GCRV) can result in large-scale death of young grass carp, Grass carp leading to irreparable economic losses that seriously affect large-scale breeding. Protein kinase C (PKC, also Grass carp reovirus (GCRV) known as PRKC) represents a family of serine/threonine protein kinases that includes multiple isozymes in many Protein kinase C (PKC) species. Among these, PKC-θ (PKC theta, also written as PRKCQ) is a novel isoform, mainly expressed in T cells, Host immune defences that is known to be involved in immune system function in mammals. To date, no research on immunological functions of fish Pkc-θ has been reported. To address this issue, we cloned the grass carp pkc-θ gene. Phylogenetic and syntenic analysis showed that this gene is the most evolutionarily conserved relative to zeb- rafish. Real-time quantitative PCR (RT-qPCR) indicated that pkc-θ was expressed at high levels in the gills and spleen of healthy grass carp. -

PKC-Theta-Mediated Signal Delivery from Thetcr/CD28 Surface Receptors
REVIEW ARTICLE published: 22 August 2012 doi: 10.3389/fimmu.2012.00273 PKC-theta-mediated signal delivery from theTCR/CD28 surface receptors Noah Isakov1* and Amnon Altman2 1 The Shraga Segal Department of Microbiology and Immunology, Faculty of Health Sciences and the Cancer Research Center, Ben-Gurion University of the Negev, Beer Sheva, Israel 2 Division of Cell Biology, La Jolla Institute for Allergy and Immunology, La Jolla, CA, USA Edited by: Protein kinase C-theta (PKCθ) is a key enzyme inT lymphocytes, where it plays an important Nick Gascoigne, Scripps Research role in signal transduction downstream of the activated T cell antigen receptor (TCR) and Institute, USA the CD28 costimulatory receptor. Interest in PKCθ as a potential drug target has increased Reviewed by: following recent findings that PKCθ is essential for harmful inflammatory responses medi- Balbino Alarcon, Consejo Superior de Investigaciones Cientificas, Spain ated byTh2 (allergies) andTh17 (autoimmunity) cells as well as for graft-versus-host disease Salvatore Valitutti, INSERM, France (GvHD) and allograft rejection, but is dispensable for beneficial responses such as antiviral *Correspondence: immunity and graft-versus-leukemia (GvL) response. TCR/CD28 engagement triggers the Noah Isakov, The Shraga Segal translocation of the cytosolic PKCθ to the plasma membrane (PM), where it localizes at Department of Microbiology and the center of the immunological synapse (IS), which forms at the contact site between an Immunology, Faculty of Health Sciences and the Cancer Research antigen-specificT cell and antigen-presenting cells (APC). However, the molecular basis for Center, Ben-Gurion University this unique localization, and whether it is required for its proper function have remained of the Negev, P.O. -
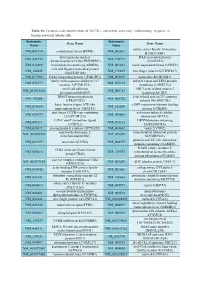
Systematic Name Gene Name Systematic Name Gene Name NM 001710 Complement Factor B(CFB) NM 052831 Solute Carrier Family 18 Member
Table S1: Genome-wide identification of SGLT2i`s interaction with early inflammatory response in human proximal tubular cells. Systematic Systematic Gene Name Gene Name Name Name solute carrier family 18 member NM_001710 complement factor B(CFB) NM_052831 B1(SLC18B1) heterogeneous nuclear DAZ associated protein NM_031372 NM_170711 ribonucleoprotein D like(HNRNPDL) 1(DAZAP1) NM_014299 bromodomain containing 4(BRD4) NM_001261 cyclin dependent kinase 9(CDK9) cilia and flagella associated protein NM_182628 NM_178835 zinc finger protein 827(ZNF827) 100(CFAP100) NM_017906 PAK1 interacting protein 1(PAK1IP1) NM_024015 homeobox B4(HOXB4) family with sequence similarity 167 ankyrin repeat and LEM domain NM_053279 NM_015114 member A(FAM167A) containing 2(ANKLE2) small cell adhesion ARP3 actin related protein 3 NM_001031628 NM_005721 glycoprotein(SMAGP) homolog(ACTR3) TRAF3 interacting protein actin related protein 2/3 complex NM_147686 NM_005720 2(TRAF3IP2) subunit 1B(ARPC1B) basic leucine zipper ATF-like cAMP responsive element binding NM_018664 NM_182898 transcription factor 3(BATF3) protein 5(CREB5) zinc finger CCCH-type containing activation induced cytidine NM_025079 NM_020661 12A(ZC3H12A) deaminase(AICDA) C-X-C motif chemokine ligand DENN domain containing NM_001511 NM_015213 1(CXCL1) 5A(DENND5A) NM_025072 prostaglandin E synthase 2(PTGES2) NM_004665 vanin 2(VNN2) superoxide dismutase 2, mitochondrial ribosomal protein NM_001024465 NM_016070 mitochondrial(SOD2) S23(MRPS23) jumonji and AT-rich interaction NM_033199 urocortin 2(UCN2) NM_004973 -

Novel Roles of SH2 and SH3 Domains in Lipid Binding
cells Review Novel Roles of SH2 and SH3 Domains in Lipid Binding Szabolcs Sipeki 1,†, Kitti Koprivanacz 2,†, Tamás Takács 2, Anita Kurilla 2, Loretta László 2, Virag Vas 2 and László Buday 1,2,* 1 Department of Molecular Biology, Institute of Biochemistry and Molecular Biology, Semmelweis University Medical School, 1094 Budapest, Hungary; [email protected] 2 Institute of Enzymology, Research Centre for Natural Sciences, 1117 Budapest, Hungary; [email protected] (K.K.); [email protected] (T.T.); [email protected] (A.K.); [email protected] (L.L.); [email protected] (V.V.) * Correspondence: [email protected] † Both authors contributed equally to this work. Abstract: Signal transduction, the ability of cells to perceive information from the surroundings and alter behavior in response, is an essential property of life. Studies on tyrosine kinase action fundamentally changed our concept of cellular regulation. The induced assembly of subcellular hubs via the recognition of local protein or lipid modifications by modular protein interactions is now a central paradigm in signaling. Such molecular interactions are mediated by specific protein interaction domains. The first such domain identified was the SH2 domain, which was postulated to be a reader capable of finding and binding protein partners displaying phosphorylated tyrosine side chains. The SH3 domain was found to be involved in the formation of stable protein sub-complexes by constitutively attaching to proline-rich surfaces on its binding partners. The SH2 and SH3 domains have thus served as the prototypes for a diverse collection of interaction domains that recognize not only proteins but also lipids, nucleic acids, and small molecules. -

Role of the Ras-Association Domain Family 1 Tumor Suppressor Gene in Human Cancers
Review Role of the Ras-Association Domain Family 1 Tumor Suppressor Gene in Human Cancers Angelo Agathanggelou, Wendy N. Cooper, and Farida Latif Section of Medical and Molecular Genetics, Division of Reproductive and Child Health, The Institute of Biomedical Research, University of Birmingham, Edgbaston, Birmingham, United Kingdom Abstract renal cell carcinomas was identified from region 3p25 confirming In recent years, the list of tumor suppressor genes (or this hypothesis (3). This prompted the search for TSGs within the candidate TSG) that are inactivated frequently by epigenetic other regions of 3p. An important TSG was suspected to reside in events rather than classic mutation/deletion events has been 3p21.3 because instability of this region is the earliest and most growing. Unlike mutational inactivation, methylation is frequently detected deficiency in lung cancer. Overlapping homo- reversible and demethylating agents and inhibitors of histone zygous deletions in lung and breast tumor cell lines reduced the deacetylases are being used in clinical trails. Highly sensitive critical region in 3p21.3 to 120 kb and this region was found to be and quantitative assays have been developed to assess exceptionally gene rich. From this critical region, eight genes were methylation in tumor samples, early lesions, and bodily fluids. identified, including CACNA2D2, PL6/placental protein 6, CYB561D2/101F6, TUSC4/NPRL2/G21, ZMYND10/BLU, RASSF1/ Hence, gene silencing by promoter hypermethylation has potential clinical benefits in early cancer diagnosis, prognosis, 123F2, TUSC2/FUS1, and HYAL2/LUCA2. However, despite extensive treatment, and prevention. The hunt for a TSG located at genetic analysis in lung and breast tumors, none of these candidate 3p21.3 resulted in the identification of the RAS-association genes were frequently mutated. -

Protein Kinase C: the “Masters” of Calcium and Lipid
Downloaded from http://cshperspectives.cshlp.org/ on September 27, 2021 - Published by Cold Spring Harbor Laboratory Press Protein Kinase C: The “Masters” of Calcium and Lipid Peter Lipp1 and Gregor Reither2 1Institute for Molecular Cell Biology, Medical Faculty, Saarland University, Homburg/Saar, Germany 2Cell Biology and Biophysics Unit, EMBL, Heidelberg, Germany Correspondence: [email protected] The coordinated and physiological behavior of living cells in an organism critically depends on their ability to interact with surrounding cells and with the extracellular space. For this, cells have to interpret incoming stimuli, correctly process the signals, and produce meaning- ful responses. A major part of such signaling mechanisms is the translation of incoming stimuli into intracellularly understandable signals, usually represented by second messen- gers or second-messenger systems. Two key second messengers, namely the calcium ion and signaling lipids, albeit extremely different in nature, play an important and often syner- gistic role in such signaling cascades. In this report, we will shed some light on an entire family of protein kinases, the protein kinases C, that are perfectly designed to exactly decode these two second messengers in all of their properties and convey the signaling content to downstream processes within the cell. nce generated, second messengers relay phosphorylation of additional signaling part- Otheir information content in a plethora ners or effector proteins. We will briefly intro- of properties, including time, quantity (i.e., duce the PKC subfamilies with particular concentration), space (i.e., subcellular distribu- emphasis on their signaling ability, discuss the tion), and interestingly into any combination of important sensing domains, and their proper- these three characteristics. -

Supplemental Data Heidel Et Al
Supplemental data Heidel et al. Table of Contents 1. Sequencing strategy and statistics ...................................................................................................... 2 2. Genome structure ............................................................................................................................... 2 2.1 Extrachromosal elements .............................................................................................................. 2 2.2 Chromosome structure ................................................................................................................. 3 2.3 Repetitive elements ...................................................................................................................... 5 3. Coding sequences ................................................................................................................................ 5 3.1 Homopolymer tracts ..................................................................................................................... 5 3.2 Gene families and orthology relationships ................................................................................... 7 3.3 Synteny analysis .......................................................................................................................... 11 4. Protein functional domains ............................................................................................................... 12 5. Protein families ................................................................................................................................. -

Identification of Novel Regulatory Mechanisms for Cdc42 Gtpase-Activating Protein Cdgap/ARHGAP31, a Protein Involved in Development and Cancer
Identification of novel regulatory mechanisms for Cdc42 GTPase-activating protein CdGAP/ARHGAP31, a protein involved in development and cancer Ali Ben Djoudi Ouadda Department of Anatomy & Cell Biology McGill University, Montréal, Québec, Canada Submitted October, 2016 A thesis submitted to McGill University in partial fulfillment of the requirements of the degree of Doctor of Philosophy © Ali Ben Djoudi Ouadda, 2016 Acknowledgments I would like to express my deepest thanks and appreciation to my supervisor, Dr Nathalie Lamarche-Vane, who has opened the door of her laboratory and gave me the opportunity to pursue an excellent research training. Without her kindness, support, guidance and persistent help this thesis would not have been possible. I would like to thank my mentor Dr. Carlos Morales, who has supported, encouraged and guided me with valuable advice from the beginning. I would like also to thank my advisory committee members, Dr. Isabelle Rouiller and Dr. Peter Siegel for their encouragement and precious scientific inputs and feedbacks. I would like to acknowledge the Fonds de Recherche du Québec-Santé (FRSQ) which awarded me a Doctoral Training Scholarship and the McGill Faculty of Medicine/Department of Anatomy & Cell Biology which granted me a Doctoral Internal Scholarship, GREAT Travel and Merit Awards. In addition, a thank you to my colleagues in the Department of Anatomy & Cell Biology, RI-MUHC, IRCM and IRIC for their help and collaboration, either with reagents or scientific discussion and troubleshooting. Special thanks to Martin, Yi and Vilayphone for their precious help and support during my early days in the lab, and to Philippe, Sadig, Fereshteh, Hidetaka, Tristan, Jonathan and Judith for their help, kindness and availability. -

Ghazi Rahman Final Phd Dis
Identification of the Activator Binding Residues in the Second Cysteine-Rich Regulatory Domain of Protein Kinase C Theta (PKCθ) A Dissertation Submitted to The Department of Pharmacological and Pharmaceutical Sciences College of Pharmacy University of Houston In Partial Fulfillment of The Requirements for the Degree of Doctor of Philosophy By Ghazi Muhammad Sayedur Rahman May 2012 “For my parents, brothers and my loving wife and their dreams” iii ACKNOWLEDGEMENTS I would like to thank my Ph.D. advisor Dr. Joydip Das for his inspiration, patience and guidance throughout the time of my dissertation. I had been through difficult times during the course of my graduate studies, which sometimes made me vulnerable, but Dr. Das has never stopped encouraging me to do the right things at right moment. He made me who I am today. As a graduate advisor he has always been methodical and perfectionist. I remember during writing the manuscript on my dissertation work, we talked a lot on what to write, how to write, what figures and tables to be incorporated and how should it be. He has been totally thorough on writing styles, on fine details of every piece of figures and illustrations and tables. He made me to change in the figures and illustrations and putting the right words in places over and over again until he finds the piece of work is absolutely perfect. Dr. Das is an excellent researcher and his passion for science is undoubtedly beyond comparison. He always told me to make one step at a time and guided me through the whole time towards the final goal. -

The C1 Domain in Cancer Signaling Molecules: Regulation by Lipids and Protein-Protein Interactions
University of Pennsylvania ScholarlyCommons Publicly Accessible Penn Dissertations Spring 2010 The C1 Domain in Cancer Signaling Molecules: Regulation by Lipids and Protein-Protein Interactions Hongbin Wang University of Pennsylvania, [email protected] Follow this and additional works at: https://repository.upenn.edu/edissertations Part of the Pharmacology, Toxicology and Environmental Health Commons Recommended Citation Wang, Hongbin, "The C1 Domain in Cancer Signaling Molecules: Regulation by Lipids and Protein-Protein Interactions" (2010). Publicly Accessible Penn Dissertations. 141. https://repository.upenn.edu/edissertations/141 This paper is posted at ScholarlyCommons. https://repository.upenn.edu/edissertations/141 For more information, please contact [email protected]. The C1 Domain in Cancer Signaling Molecules: Regulation by Lipids and Protein- Protein Interactions Abstract Cysteine-rich (C1) domains, present in PKC isozymes, Chimaerins, RasGRPs, PKDs, Munc13s, DGKs, and MRCKs, can bind the diacylglycerol (DAG) second messenger. In the present thesis research, I demonstrated that p23/Tmp21 acts as a C1-domain docking protein that mediates perinuclear translocation of beta2-chimaerin. Glu227 and Leu248 in the beta2-chimaerin C1-domain are crucial for binding p23/Tmp21 and perinuclear targeting. Isolated C1-domains from individual PKC isozymes or RasGRP1 differentially interact with p23/Tmp21. PKCepsilon interacts with p23/Tmp21 specifically via its C1b domain, however this association is lost in response to phorbol esters. These results demonstrate that p23/Tmp21 acts as an anchor that distinctively modulates compartmentalization of C1-domain- containing proteins, and it plays an essential role in beta2-chimaerin re-localization to the perinuclear region in response to phorbol esters. It has been established that apoptosis induced by phorbol esters in LNCaP cells is primarily mediated by the novel PKCdelta. -

A) Analysis of the Ncrnas of Arabidopsis Inflorescences and Pollen Bioreplicates (Two Bioreplicates for Each Tissue
Supplementary Material: Supplemental Figure 1. (A) Analysis of the ncRNAs of Arabidopsis inflorescences and pollen bioreplicates (two bioreplicates for each tissue). Biological replicates 1 and 2 of inflorescence are paired samples for both pollen biological replicates 1 and 2. (B) Individual tRF accumulation profiles for the different sRNA libraries used in Figure 1C. (C-D) Distribution of 5’ end nucleotide of 19 nt sRNAs along mature tRNA transcript sequences post- transcriptionally modified with the CCA sequence, in inflorescence (C) and pollen (D). Supplemental Figure 2. (A-C) Distribution of 5’ end nucleotide of 19 nt tRFs along mature tRNA transcript sequences post-transcriptionally modified with the CCA sequence in rice (A) and maize (B) pollen, and Physcomitrella patens gametophote-sporophyte (C) samples. Supplemental Figure 3. (A) Individual tRF accumulation profiles for the different sRNA libraries used in Figure 3A. (B) tRF accumulation profile in seedling sRNA libraries from ddm1 (red) and wt (blue), normalized in RPM. (C) Distribution of 19 nt tRFs along mature tRNA transcript sequences post- transcriptionally modified with the CCA sequence in the ddm1 background. (D) tRF accumulation profile in inflorescence sRNA libraries from met1 single and ddc triple mutants. 1 Supplemental Figure 4. (A) Individual tRF accumulation profiles for the different sRNA libraries used in Figure 4B. (B) AGO1-immunoprecipitated tRF accumulation size profile in wt and ddm1. (C) NcRNA categorization of the AGO1-immunoprecipitated sRNAs in wt and ddm1 libraries. (D) RT-PCR analysis of the accumulation of tRNA transcripts in gene-specific reverse (1) or oligo-dT (2) primers synthesized cDNA for selected tRNAs in the ddm1, ddm1/dcl1-11 and ddm1/ago1-24 backgrounds. -

Viewed and Counted at the Microscope Or Images Were Obtained
A ThesisThesis entitled Identification and characterization of RhoGAPs involved in the regulation of invadopodia by Kyle Lee Snyder Submitted to the Graduate Faculty as partial fulfillment of the requirements for the Master of Science Degree in Cellular and Molecular Biology _________________________________________ Dr. Rafael Garcia-Mata, Committee Chair _________________________________________ Dr. Deborah Chadee, Committee Member _________________________________________ Dr. Song-Tao Liu, Committee Member _________________________________________ Dr. Patricia R. Komuniecki, Dean College of Graduate Studies The University of Toledo April, 2016 Copyright 2016, Kyle Lee Snyder This document is copyrighted material. Under copyright law, no parts of this document may be reproduced without the expressed permission of the author. An Abstract of Identification and characterization of RhoGAPs involved in the regulation of invadopodia by Kyle Lee Snyder Submitted to the Graduate Faculty as partial fulfillment of the requirements for the Master of Science Degree in Cellular and Molecular Biology The University of Toledo April, 2016 Invadopodia are actin rich structures that enhance a cancer cells ability to degrade the extracellular matrix (ECM) and promote metastasis. Formation of invadopodia is regulated by Rho GTPases, a family of small G proteins that regulate actin rearrangement, cellular migration, and invasion. These proteins exist in two states, inactive GDP-bound, and active GTP-bound conformations. Activation is regulated by GEFs (guanine nucleotide exchange factors), whereas inactivation is modulated by GAPs (GTPase activating proteins). In our preliminary studies we screened 18 members of the RhoGAP family to identify if any were involved in signaling events contributing to invadopodia formation. We identified three candidates, TCGAP, CHN1 and ARHGAP12. We have confirmed that the knockdown of each of these genes is sufficient to increase invadopodia formation, and validated these results with over expression studies.