Pearson Correlation Analysis Was Used to Assess the Correlation Between Samples by Using the R Version 3.6.3
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
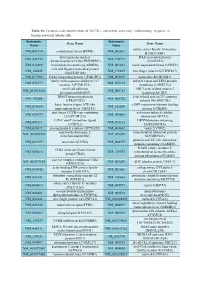
Systematic Name Gene Name Systematic Name Gene Name NM 001710 Complement Factor B(CFB) NM 052831 Solute Carrier Family 18 Member
Table S1: Genome-wide identification of SGLT2i`s interaction with early inflammatory response in human proximal tubular cells. Systematic Systematic Gene Name Gene Name Name Name solute carrier family 18 member NM_001710 complement factor B(CFB) NM_052831 B1(SLC18B1) heterogeneous nuclear DAZ associated protein NM_031372 NM_170711 ribonucleoprotein D like(HNRNPDL) 1(DAZAP1) NM_014299 bromodomain containing 4(BRD4) NM_001261 cyclin dependent kinase 9(CDK9) cilia and flagella associated protein NM_182628 NM_178835 zinc finger protein 827(ZNF827) 100(CFAP100) NM_017906 PAK1 interacting protein 1(PAK1IP1) NM_024015 homeobox B4(HOXB4) family with sequence similarity 167 ankyrin repeat and LEM domain NM_053279 NM_015114 member A(FAM167A) containing 2(ANKLE2) small cell adhesion ARP3 actin related protein 3 NM_001031628 NM_005721 glycoprotein(SMAGP) homolog(ACTR3) TRAF3 interacting protein actin related protein 2/3 complex NM_147686 NM_005720 2(TRAF3IP2) subunit 1B(ARPC1B) basic leucine zipper ATF-like cAMP responsive element binding NM_018664 NM_182898 transcription factor 3(BATF3) protein 5(CREB5) zinc finger CCCH-type containing activation induced cytidine NM_025079 NM_020661 12A(ZC3H12A) deaminase(AICDA) C-X-C motif chemokine ligand DENN domain containing NM_001511 NM_015213 1(CXCL1) 5A(DENND5A) NM_025072 prostaglandin E synthase 2(PTGES2) NM_004665 vanin 2(VNN2) superoxide dismutase 2, mitochondrial ribosomal protein NM_001024465 NM_016070 mitochondrial(SOD2) S23(MRPS23) jumonji and AT-rich interaction NM_033199 urocortin 2(UCN2) NM_004973 -

Supplemental Data Heidel Et Al
Supplemental data Heidel et al. Table of Contents 1. Sequencing strategy and statistics ...................................................................................................... 2 2. Genome structure ............................................................................................................................... 2 2.1 Extrachromosal elements .............................................................................................................. 2 2.2 Chromosome structure ................................................................................................................. 3 2.3 Repetitive elements ...................................................................................................................... 5 3. Coding sequences ................................................................................................................................ 5 3.1 Homopolymer tracts ..................................................................................................................... 5 3.2 Gene families and orthology relationships ................................................................................... 7 3.3 Synteny analysis .......................................................................................................................... 11 4. Protein functional domains ............................................................................................................... 12 5. Protein families ................................................................................................................................. -

Identification of Novel Regulatory Mechanisms for Cdc42 Gtpase-Activating Protein Cdgap/ARHGAP31, a Protein Involved in Development and Cancer
Identification of novel regulatory mechanisms for Cdc42 GTPase-activating protein CdGAP/ARHGAP31, a protein involved in development and cancer Ali Ben Djoudi Ouadda Department of Anatomy & Cell Biology McGill University, Montréal, Québec, Canada Submitted October, 2016 A thesis submitted to McGill University in partial fulfillment of the requirements of the degree of Doctor of Philosophy © Ali Ben Djoudi Ouadda, 2016 Acknowledgments I would like to express my deepest thanks and appreciation to my supervisor, Dr Nathalie Lamarche-Vane, who has opened the door of her laboratory and gave me the opportunity to pursue an excellent research training. Without her kindness, support, guidance and persistent help this thesis would not have been possible. I would like to thank my mentor Dr. Carlos Morales, who has supported, encouraged and guided me with valuable advice from the beginning. I would like also to thank my advisory committee members, Dr. Isabelle Rouiller and Dr. Peter Siegel for their encouragement and precious scientific inputs and feedbacks. I would like to acknowledge the Fonds de Recherche du Québec-Santé (FRSQ) which awarded me a Doctoral Training Scholarship and the McGill Faculty of Medicine/Department of Anatomy & Cell Biology which granted me a Doctoral Internal Scholarship, GREAT Travel and Merit Awards. In addition, a thank you to my colleagues in the Department of Anatomy & Cell Biology, RI-MUHC, IRCM and IRIC for their help and collaboration, either with reagents or scientific discussion and troubleshooting. Special thanks to Martin, Yi and Vilayphone for their precious help and support during my early days in the lab, and to Philippe, Sadig, Fereshteh, Hidetaka, Tristan, Jonathan and Judith for their help, kindness and availability. -

Viewed and Counted at the Microscope Or Images Were Obtained
A ThesisThesis entitled Identification and characterization of RhoGAPs involved in the regulation of invadopodia by Kyle Lee Snyder Submitted to the Graduate Faculty as partial fulfillment of the requirements for the Master of Science Degree in Cellular and Molecular Biology _________________________________________ Dr. Rafael Garcia-Mata, Committee Chair _________________________________________ Dr. Deborah Chadee, Committee Member _________________________________________ Dr. Song-Tao Liu, Committee Member _________________________________________ Dr. Patricia R. Komuniecki, Dean College of Graduate Studies The University of Toledo April, 2016 Copyright 2016, Kyle Lee Snyder This document is copyrighted material. Under copyright law, no parts of this document may be reproduced without the expressed permission of the author. An Abstract of Identification and characterization of RhoGAPs involved in the regulation of invadopodia by Kyle Lee Snyder Submitted to the Graduate Faculty as partial fulfillment of the requirements for the Master of Science Degree in Cellular and Molecular Biology The University of Toledo April, 2016 Invadopodia are actin rich structures that enhance a cancer cells ability to degrade the extracellular matrix (ECM) and promote metastasis. Formation of invadopodia is regulated by Rho GTPases, a family of small G proteins that regulate actin rearrangement, cellular migration, and invasion. These proteins exist in two states, inactive GDP-bound, and active GTP-bound conformations. Activation is regulated by GEFs (guanine nucleotide exchange factors), whereas inactivation is modulated by GAPs (GTPase activating proteins). In our preliminary studies we screened 18 members of the RhoGAP family to identify if any were involved in signaling events contributing to invadopodia formation. We identified three candidates, TCGAP, CHN1 and ARHGAP12. We have confirmed that the knockdown of each of these genes is sufficient to increase invadopodia formation, and validated these results with over expression studies. -

ARHGAP4 IS a SPATIALLY REGULATED RHOGAP THAT INHIBITS NIH/3T3 CELL MIGRATION and DENTATE GRANULE CELL AXON OUTGROWTH by DANIEL L
ARHGAP4 IS A SPATIALLY REGULATED RHOGAP THAT INHIBITS NIH/3T3 CELL MIGRATION AND DENTATE GRANULE CELL AXON OUTGROWTH By DANIEL LEE VOGT Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy Department of Neuroscience CASE WESTERN RESERVE UNIVERSITY August, 2007 CASE WESTERN RESERVE UNIVERSITY SCHOOL OF GRADUATE STUDIES We hereby approve the dissertation of Daniel Lee Vogt ______________________________________________________ candidate for the Ph.D. degree *. (signed) (chair of the committee)________________________________ Stefan Herlitze ________________________________________________Alfred Malouf Robert Miller ________________________________________________ ________________________________________________Thomas Egelhoff ________________________________________________Susann Brady-Kalnay ________________________________________________ (date) _______________________6-21-2007 *We also certify that written approval has been obtained for any proprietary material contained therein. ii Copyright © 2007 by Daniel Lee Vogt All rights reserved iii Table of contents Page # Title page i Table of contents iv List of figures vii Abstract 1 Chapter one: General introduction 2 Hippocampal axon pathways and development 3 Guidance cues in hippocampal axon outgrowth 6 Slit/Robo 7 Semaphorins, plexins and neuropilins 8 Ephrins and ephs 11 Other guidance cues in the hippocampus 13 GTPases: structure and function of ras superfamily members 15 Ras GTPases 17 Ran GTPases 18 Arf GTPases 18 Rab GTPases 19 Rho GTPases -

Functional Analysis of a Rho Gtpase Activating Protein Involved In
Functional Analysis of a Rho GTPase Activating Protein Involved in Epithelial Differentiation and Morphogenesis Ahmed Nehad Elbediwy Thesis submitted to University College London for the award of Doctor of Philosophy November 2012 Department of Cell Biology Institute of Ophthalmology University College London 11-43 Bath Street London EC1V 9EL Declaration of Work I, Ahmed Nehad Elbediwy, hereby confirm that the work that is presented in this thesis is my own. Where information has been derived from alternate sources, I confirm that I have indicated this in the correct context within the thesis University College London November 2012 2 Abstract Polarized epithelial cells form selective barriers between tissues and various body compartments that are essential for normal development and organ function. A mature apical junctional complex (AJC), consisting of tight junctions (TJ), adherens junctions (AJ), and desmosomes is crucial for functional polarized epithelia. Rho-GTPases are key regulatory proteins of many cellular processes, including epithelial adhesion and polarization. These small GTPases are in turn controlled spatially and temporally by guanine nucleotide exchange factors (GEFs) that promote GTP-binding, resulting in their activation; and GTPase activating proteins (GAPs) that promote GDP hydrolysis, resulting in their inactivation. In this thesis I studied a novel junction associated GAP protein known as SH3BP1 in a variety of epithelia. SH3BP1 was identified in a functional siRNA screen that was designed to identify actin regulators of epithelial polarisation and differentiation. I will show that SH3BP1 localises to the early AJC when TJ and AJ are not yet properly separated. SH3BP1 regulation is important for tight junction formation and, its depletion affects polarity and junction integrity. -

Roots of Angiosperm Formins: the Evolutionary History of Plant FH2 Domain-Containing Proteins Michal Grunt1, Viktor Žárský1,2 and Fatima Cvrčková*1
BMC Evolutionary Biology BioMed Central Research article Open Access Roots of angiosperm formins: The evolutionary history of plant FH2 domain-containing proteins Michal Grunt1, Viktor Žárský1,2 and Fatima Cvrčková*1 Address: 1Department of Plant Physiology, Faculty of Sciences, Charles University, Vinièná 5, CZ 128 43 Praha 2, Czech Republic and 2Institute of Experimental Botany, Academy of Sciences of the Czech Republic, Rozvojová 135, CZ 165 02 Praha 6, Czech Republic Email: Michal Grunt - [email protected]; Viktor Žárský - [email protected]; Fatima Cvrčková* - [email protected] * Corresponding author Published: 22 April 2008 Received: 19 December 2007 Accepted: 22 April 2008 BMC Evolutionary Biology 2008, 8:115 doi:10.1186/1471-2148-8-115 This article is available from: http://www.biomedcentral.com/1471-2148/8/115 © 2008 Grunt et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: Shuffling of modular protein domains is an important source of evolutionary innovation. Formins are a family of actin-organizing proteins that share a conserved FH2 domain but their overall domain architecture differs dramatically between opisthokonts (metazoans and fungi) and plants. We performed a phylogenomic analysis of formins in most eukaryotic kingdoms, aiming to reconstruct an evolutionary scenario that may have produced the current diversity of domain combinations with focus on the origin of the angiosperm formin architectures. Results: The Rho GTPase-binding domain (GBD/FH3) reported from opisthokont and Dictyostelium formins was found in all lineages except plants, suggesting its ancestral character. -

Phylogenetic Analysis of Rhogap Domain-Containing Proteins
Letter Phylogenetic Analysis of RhoGAP Domain-Containing Proteins Marcel0 M. Brand&, Karina L. Silva-Brandh2, Fernando F. Costal, and Sara T.O. Saadl* Hematology and Hemotherapy Center, State University of Campinas, 13083-970 Campinas, SP, Brazil; Department of Zoology, Institute of Biology, State University of Campinas, 13083-970 Campinas, SP, Brazil. Proteins containing an Rho GTPase-activating protein (RhoGAP) domain work as molecular switches involved in the regulation of diverse cellular functions. The abil- ity of these GTPases to regulate a wide number of cellular processes comes from their interactions with multiple effectors and inhibitors, including the RhoGAP family, which stimulates their intrinsic GTPase activity. Here, a phylogenetic approach was applied to study the evolutionary relationship among 59 RhoGAP domain-containing proteins. The sequences were aligned by their RhoGAP do- mains and the phylogenetic hypotheses were generated using Maximum Parsimony and Bayesian analyses. The character tracing of two traits, GTPase activity and presence of other domains, indicated a significant phylogenetic signal for both of them. words: Bayesian analysis, character tracing, parsimony, phylogenomics, protein domain, RhoGAP Introduction The Rho GTPase-activating proteins (RhoGAPs) are of GTP hydrolysis. defined by the presence of a 150-amino-acid homolog In the early analyses of the human genome se- region that is designated as the RhoGAP domain. quence, 77 different genes containing the RhoGAP This domain is necessary and sufficient for GAP domain were found. Further studies have demon- activity and shares at least 20% sequence identity strated that many of these genes are simple gene se- among its family members (1,2). Proteins contain- quence variations or single nucleotide polymorphisms ing an RhoGAP domain act as molecular switches in- (6,7). -

High Resolution Genome Wide Expression Analysis of Single Myofibers Using SMART-Seq
bioRxiv preprint doi: https://doi.org/10.1101/724393; this version posted October 17, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Blackburn et al. 2019 Manuscript High Resolution Genome Wide Expression Analysis of Single Myofibers Using SMART-Seq Darren M. Blackburn1,2,*, Felicia Lazure1,2,*, Aldo H. Corchado1, Theodore J. Perkins3,4, Hamed S. Najafabadi1, Vahab D. Soleimani1,2,5 1Department of Human Genetics, McGill University, 3640 rue University, Montreal, QC, H3A 0C7 Canada 2Molecular and Regenerative Medicine Axis, Lady Davis Institute for Medical ResearcH, JewisH General Hospital, 3755 Chemin de la Cote-Sainte-CatHerine, Montreal, QC, H3T 1E2, Canada 3Sprott Center for Stem Cell Research, Ottawa Hospital Research Institute, 501 Smyth Road, Ottawa, ON, K1H 8L6, Canada 4Department of BiocHemistry, Microbiology and Immunology, University of Ottawa, 451 SmytH Road, Ottawa, ON, K1H 8M5, Canada 5To whom correspondence sHould be addressed: VaHab D. Soleimani: Department of Human Genetics, McGill University, 3640 rue University, Montreal, QC, H3A 0C7, Canada; [email protected]; TelepHone number: +1 514 340 8222 ext. 26136; Fax number: +1 514 340-7502. *These autHors made equal contributions Running title: Single myofiber RNA-Seq Key Words: Single Myofiber, Skeletal Muscle, SMART-Seq, Gene Expression Analysis, Myofiber Heterogeneity, Aging skeletal muscle Word count: 3930 1 bioRxiv preprint doi: https://doi.org/10.1101/724393; this version posted October 17, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved.