The Mop1 Mutation Affects the Recombination Landscape in Maize
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Framework for In-Field Analyses of Performance and Sub-Technique Selection in Standing Para Cross-Country Skiers
sensors Article Framework for In-Field Analyses of Performance and Sub-Technique Selection in Standing Para Cross-Country Skiers Camilla H. Carlsen 1,*, Julia Kathrin Baumgart 1, Jan Kocbach 1,2, Pål Haugnes 1 , Evy M. B. Paulussen 1,3 and Øyvind Sandbakk 1 1 Centre for Elite Sports Research, Department of Neuromedicine and Movement Science, Faculty of Medicine and Health Sciences, Norwegian University of Science and Technology, 7491 Trondheim, Norway; [email protected] (J.K.B.); [email protected] (J.K.); [email protected] (P.H.); [email protected] (E.M.B.P.); [email protected] (Ø.S.) 2 NORCE Norwegian Research Centre AS, 5008 Bergen, Norway 3 Faculty of Health, Medicine & Life Sciences, Maastricht University, 6200 MD Maastricht, The Netherlands * Correspondence: [email protected]; Tel.: +47-452-40-788 Abstract: Our aims were to evaluate the feasibility of a framework based on micro-sensor technology for in-field analyses of performance and sub-technique selection in Para cross-country (XC) skiing by using it to compare these parameters between elite standing Para (two men; one woman) and able- bodied (AB) (three men; four women) XC skiers during a classical skiing race. The data from a global navigation satellite system and inertial measurement unit were integrated to compare time loss and selected sub-techniques as a function of speed. Compared to male/female AB skiers, male/female Para skiers displayed 19/14% slower average speed with the largest time loss (65 ± 36/35 ± 6 s/lap) Citation: Carlsen, C.H.; Kathrin found in uphill terrain. -

Table S1. Cryo-EM Data Acquisition and Image Processing
Table S1. Cryo-EM data acquisition and image processing. Data Collection Electron Microscope Titan Krios Camera Falcon 4 Voltage (kV) 300 Nominal Magnification 75,000 Calibrated physical pixel size (Å) 1.03 Total exposure (e/Å2) 42 Exposure rate (e/pixel/s) 4.6 Number of frames 29 Defocus range (μm) 0.9 to 2 Image Processing Motion correction software MotionCor2 CTF estimation software cryoSPARC v2 Particle selection software cryoSPARC v2 Micrographs used in inhibitor free dataset 3,953 Micrographs used in Inz-5 bound dataset 4,396 Particle images selected in inhibitor free 1,151,426 dataset Particle images selected in Inz-5 bound dataset 1,129,413 3D map classification and refinement software cryoSPARC v2 Table S2. CryoEM map and atomic model statistics. Dataset Inhibitor Inz-5 free bound Associated PDB ID Modelling and Coot, Coot, refinement phenix, phenix, software ISOLDE ISOLDE Protein residues 1906 1909 Ligand HEM:3, HEM:3, FES:1, FES:1, UQ:2 INZ:1 RMSD bond length 0.004 0.003 (Å) RMSD bond angle 0.677 0.691 (°) Ramachandran 0 0 outliers (%) Ramachandran 96.13 97.25 favoured (%) Rotamer outliers 0 0 (%) Clash score 8.92 8.97 MolProbability 1.74 1.62 score EMringer score 4.25 4.04 Fig. S1. CryoEM map calculations for inhibitor free dataset. (A) UV-visible spectra of purified CIII2. (B) Representative micrograph and 2D class average images for CIII2. Scale bar, 500 Å. (C) Fourier shell correlation (FSC) curve after correction for solvent masking. (D) Viewing direction distribution for particle images. (E) Local resolution estimate for CIII2 map from non-uniform refinement with C2 symmetry. -

GUIDE to ADAPTED SWIMMING CLASSIFICATIONS Swimming Is
GUIDE TO ADAPTED SWIMMING CLASSIFICATIONS Swimming is the only sport that combines the conditions of limb loss, cerebral palsy (coordination and movement restrictions), spinal cord injury (weakness or paralysis involving any combination of the limbs) and other disabilities (such as Dwarfism (little people); major joint restriction conditions) across classes. Classes 1-10 – are allocated to swimmers with a physical disability Classes 11-13 – are allocated to swimmers with a visual disability Class 14 – is allocated to swimmers with an intellectual disability The Prefix S to the Class denotes the class for Freestyle, Backstroke and Butterfly The Prefix SB to the class denotes the class for Breaststroke The Prefix SM to the class denotes the class for Individual Medley. The range is from the swimmers with severe disability (S1, SB1, SM1) to those with the minimal disability (S10, SB9, SM10) In any one class some swimmers may start with a dive or in the water depending on their condition. This is factored in when classifying the athlete. The examples are only a guide – some conditions not mentioned may also fit the following classes. Locomotor Impaired (S1-S10): S1: Generally persons with complete spinal cord injuries below C4-C5 or cerebral palsy characterized by severe quadriplegia. Unable to catch the water. Severely limited propulsion from the arms due to muscle weakness, restricted range of motion or uncoordinated movements. No trunk control. No functional leg movements and significant leg drag. Assisted water start. Ordinarily uses the backstroke because of an inability to turn the head to breathe when swimming freestyle. S2: Generally persons with complete spinal cord injuries below C6-C7 or similar musculoskeletal impairment or cerebral palsy characterized by severe quadriplegia. -

National Classification? 13
NATIONAL CL ASSIFICATION INFORMATION FOR MULTI CLASS SWIMMERS Version 1.2 2019 PRINCIPAL PARTNER MAJOR PARTNERS CLASSIFICATION PARTNERS Version 1.2 2019 National Swimming Classification Information for Multi Class Swimmers 1 CONTENTS TERMINOLOGY 3 WHAT IS CLASSIFICATION? 4 WHAT IS THE CLASSIFICATION PATHWAY? 4 WHAT ARE THE ELIGIBLE IMPAIRMENTS? 5 CLASSIFICATION SYSTEMS 6 CLASSIFICATION SYSTEM PARTNERS 6 WHAT IS A SPORT CLASS? 7 HOW IS A SPORT CLASS ALLOCATED TO AN ATHLETE? 7 WHAT ARE THE SPORT CLASSES IN MULTI CLASS SWIMMING? 8 SPORT CLASS STATUS 11 CODES OF EXCEPTION 12 HOW DO I CHECK MY NATIONAL CLASSIFICATION? 13 HOW DO I GET A NATIONAL CLASSIFICATION? 13 MORE INFORMATION 14 CONTACT INFORMATION 16 Version 1.2 2019 National Swimming Classification Information for Multi Class Swimmers 2 TERMINOLOGY Assessment Specific clinical procedure conducted during athlete evaluation processes ATG Australian Transplant Games SIA Sport Inclusion Australia BME Benchmark Event CISD The International Committee of Sports for the Deaf Classification Refers to the system of grouping athletes based on impact of impairment Classification Organisations with a responsibility for administering the swimming classification systems in System Partners Australia Deaflympian Representative at Deaflympic Games DPE Daily Performance Environment DSA Deaf Sports Australia Eligibility Criteria Requirements under which athletes are evaluated for a Sport Class Evaluation Process of determining if an athlete meets eligibility criteria for a Sport Class HI Hearing Impairment ICDS International Committee of Sports for the Deaf II Intellectual Impairment Inas International Federation for Sport for Para-athletes with an Intellectual Disability General term that refers to strategic initiatives that address engagement of targeted population Inclusion groups that typically face disadvantage, including people with disability. -

(VA) Veteran Monthly Assistance Allowance for Disabled Veterans
Revised May 23, 2019 U.S. Department of Veterans Affairs (VA) Veteran Monthly Assistance Allowance for Disabled Veterans Training in Paralympic and Olympic Sports Program (VMAA) In partnership with the United States Olympic Committee and other Olympic and Paralympic entities within the United States, VA supports eligible service and non-service-connected military Veterans in their efforts to represent the USA at the Paralympic Games, Olympic Games and other international sport competitions. The VA Office of National Veterans Sports Programs & Special Events provides a monthly assistance allowance for disabled Veterans training in Paralympic sports, as well as certain disabled Veterans selected for or competing with the national Olympic Team, as authorized by 38 U.S.C. 322(d) and Section 703 of the Veterans’ Benefits Improvement Act of 2008. Through the program, VA will pay a monthly allowance to a Veteran with either a service-connected or non-service-connected disability if the Veteran meets the minimum military standards or higher (i.e. Emerging Athlete or National Team) in his or her respective Paralympic sport at a recognized competition. In addition to making the VMAA standard, an athlete must also be nationally or internationally classified by his or her respective Paralympic sport federation as eligible for Paralympic competition. VA will also pay a monthly allowance to a Veteran with a service-connected disability rated 30 percent or greater by VA who is selected for a national Olympic Team for any month in which the Veteran is competing in any event sanctioned by the National Governing Bodies of the Olympic Sport in the United State, in accordance with P.L. -

A Strategy to Optimize the Generation of Stable Chromobody Cell Lines for Visualization and Quantification of Endogenous Proteins in Living Cells
Supplementary: A strategy to optimize the generation of stable chromobody cell lines for visualization and quantification of endogenous proteins in living cells Bettina-Maria Keller1, Julia Maier1, Melissa Weldle1, Soeren Segan2, Bjoern Traenkle1, Ulrich Rothbauer1,2* 1 Pharmaceutical Biotechnology, Eberhard Karls University Tuebingen, Germany 2 Natural and Medical Sciences Institute at the University of Tuebingen, Reutlingen, Germany Correspondence: Prof. Dr. Ulrich Rothbauer, Natural and Medical Sciences Institute at the University of Tuebingen Markwiesenstr. 55, 72770 Reutlingen, Germany. E-mail: [email protected] Phone: +49 7121 51530-415 Fax: +49 7121 51530-816 Supplementary information DNA oligo name Sequence, 5’ - 3’ NB-ubi-for ATA TAT CTG CAG GAG TCT GGG GGA GGC TTG GTG CA NB-ubi-rev ATA TAT TCC GGA GGA GAC GGT GAC CTG GGT CCC β-actin-promoter-for GGA ATT AAT ACT GCC TGG CCA CTC CAT G β-actin-promoter-rev TCC GCT AGC TCG GCA AAG GCG AGG C β-actin-promoter-mutPstI-for AGA GCT CCT TGT GCA GGA GCG β-actin-promoter-mutPstI-rev TGG AGG GCA TGG AGT GGC AAVS1-CB-donor-fragment-2-for TAG AGG CGG CAA TTG TTC A AAVS1-CB-donor-fragment-2-rev TGT TGT TAA CTT GTT TAT TGC AGC Seq-AAVS1-CB-donor-1 TAT GGA AAA ACG CCA GCA AC Seq-AAVS1-CB-donor-2 ATG TGG CTC TGG TTC TGG G Seq-AAVS1-CB-donor-3 AGC GGC TCG GCT TCA C Seq-AAVS1-CB-donor-4 CCT TAG ATG TTT TAC TAG CCA GAT genPCR-AAVS1-int-for TCG ACT TCC CTT CTT CCG ATG genPCR-AAVS1-int-rev CTC AGA TTC TGG GAG AGG GTA EF1α-promoter (gBlock® gene TTACCGCCATGCATTAGTTATTAATGGCTCCGGTGCCCGTCAGTGGGCAGAGCGCACAT -

The Paralympic Athlete Dedicated to the Memory of Trevor Williams Who Inspired the Editors in 1997 to Write This Book
This page intentionally left blank Handbook of Sports Medicine and Science The Paralympic Athlete Dedicated to the memory of Trevor Williams who inspired the editors in 1997 to write this book. Handbook of Sports Medicine and Science The Paralympic Athlete AN IOC MEDICAL COMMISSION PUBLICATION EDITED BY Yves C. Vanlandewijck PhD, PT Full professor at the Katholieke Universiteit Leuven Faculty of Kinesiology and Rehabilitation Sciences Department of Rehabilitation Sciences Leuven, Belgium Walter R. Thompson PhD Regents Professor Kinesiology and Health (College of Education) Nutrition (College of Health and Human Sciences) Georgia State University Atlanta, GA USA This edition fi rst published 2011 © 2011 International Olympic Committee Blackwell Publishing was acquired by John Wiley & Sons in February 2007. Blackwell’s publishing program has been merged with Wiley’s global Scientifi c, Technical and Medical business to form Wiley-Blackwell. Registered offi ce: John Wiley & Sons, Ltd, The Atrium, Southern Gate, Chichester, West Sussex, PO19 8SQ, UK Editorial offi ces: 9600 Garsington Road, Oxford, OX4 2DQ, UK The Atrium, Southern Gate, Chichester, West Sussex, PO19 8SQ, UK 111 River Street, Hoboken, NJ 07030-5774, USA For details of our global editorial offi ces, for customer services and for information about how to apply for permission to reuse the copyright material in this book please see our website at www.wiley.com/wiley-blackwell The right of the author to be identifi ed as the author of this work has been asserted in accordance with the UK Copyright, Designs and Patents Act 1988. All rights reserved. No part of this publication may be reproduced, stored in a retrieval system, or transmitted, in any form or by any means, electronic, mechanical, photocopying, recording or otherwise, except as permitted by the UK Copyright, Designs and Patents Act 1988, without the prior permission of the publisher. -

Athletes with Disability Handbook 2009
1 Athletes with Disability Handbook 2009 Athletes with Disability Handbook ATHLETES WITH DISABILITY COMMITTEE Canadian Academy of Sport Medicine 5330 rue Canotek Road, Unit (é) 4 Ottawa, (ON) K1J 9C1 Tel. (613) 748-5851 Fax (613 748-5792 1-877-585-2394 Internet: [email protected] www.casm-acms.org 2 Athletes with Disability Handbook 2009 Acknowledgements: A special thanks to Dr. Dhiren Naidu, Dr. Nancy Dudek, and Dr. Doug Dittmer for helping with the organization and content of this manual. I would also like to thank the many authors who contributed their time and expertise to write the chapters in this manual. Without your help this project would not have been a success. Sincerely, Russ O’Connor MD, FRCPC, CASM Dip Sport Med 3 Athletes with Disability Handbook 2009 Table of Contents 1. RED FLAGS .................................................................................................................. 5 TOPIC: CHANGE IN MOTOR OR SENSORY FUNCTION ......................................................... 6 TOPIC: NEW OR SIGNIFICANT CHANGE IN SPASTICITY ...................................................... 7 TOPIC: AUTONOMIC DYSREFLEXIA (AD) .................................................................................. 8 TOPIC: FRACTURES IN A PARALYZED ATHLETE ................................................................... 9 TOPIC: SWOLLEN LIMB IN AN ATHLETE WITH A NEUROLOGICAL IMPAIRMENT ....... 10 TOPIC: BALCOFEN WITHDRAWAL SYNDROME (BWS) ....................................................... 11 TOPIC: FEVER ................................................................................................................................ -
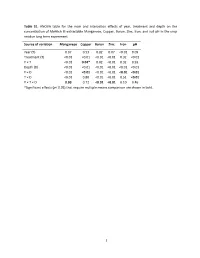
Table S1. ANOVA Table for the Main and Interaction Effects of Year
Table S1. ANOVA table for the main and interaction effects of year, treatment and depth on the concentration of Mehlich III extractable Manganese, Copper, Boron, Zinc, Iron, and soil pH in the crop residue long term experiment. Source of variation Manganese Copper Boron Zinc Iron pH Year (Y) 0.07 0.53 0.02 0.07 <0.01 0.09 Treatment (T) <0.01 <0.01 <0.01 <0.01 0.32 <0.01 Y × T <0.01 0.04* 0.02 <0.01 0.32 0.93 Depth (D) <0.01 <0.01 <0.01 <0.01 <0.01 <0.01 Y × D <0.01 <0.01 <0.01 <0.01 <0.01 <0.01 T × D <0.01 0.80 <0.01 <0.01 0.51 <0.01 Y × T × D 0.03 0.72 <0.01 <0.01 0.10 0.46 *Significant effects (p< 0.05) that require multiple means comparison are shown in bold. 1 Table S2. Mean concentration of Mehlich III extractable manganese obtained from the 120 combinations of Treatment, Year and Depth (cm) after 84 years for the crop residue long-term experiment. Depth (cm) FB1 GP FYM NB NB45 NB90 PV SB SB45 SB90 (mg Mn kg-1) 1995 0-10 B2 102.1 ab3 A 176.5 a A 163.4 a B 116.8 a B 107.8 a B 105.5 ab A 150.5 a B 111.9 a B 93.2 a B 101.6 a 10-20 CD 108.0 a A 148.7 a AB 143.5 a CD 111.8 a D 103.3 a CD 109.2 a ABC 133.3 a BCD 115.1 a D 95.7 a D 97.9 a 20-30 ABC 78.3 bc AB 90.3 bcd ABC 80.2 cd ABC 77.8 b BC 75.0 bc BC 70.9 cd A 100.8 b AB 85.4 b C 64.0 bcd BC 66.0 bc 30-60 ABC 66.6 cd A 89.4 bcd AB 76.7 cd C 55.5 cd BC 63.9 bcd BC 61.4 cde AB 82.6 bc BC 61.8 cde C 48.1 de C 48.9 cd 2005 0-10 B 97.5 ab A 149.1 a B 96.5 bc B 88.6 ab B 83.3 ab B 81.6 bc B 78.7 bc B 85.2 b B 82.1 ab B 80.2 ab 10-20 A 61.0 cde A 79.5 cd A 80.1 cd A 72.0 bc A 68.7 bc A 71.5 -

Supporting Information
Supporting Information Direct Synthesis of Bulk Boron-Doped Graphitic Carbon Nicholas P. Stadie, Emanuel Billeter, Laura Piveteau, Kostiantyn V. Kravchyk, Max Döbeli, and Maksym V. Kovalenko Contents: Tables S1-S3. Acquisition parameters for 11B and 13C NMR experiments Figure S1. Photographs of precursors and BC3′ product 13 Figure S2. C MAS NMR spectrum of BC3′ Figure S3. Raman spectroscopy of reference materials Figure S4. XRD comparison between tiled BC3′ and BC3′ (this work) Figure S5. SEM comparison between tiled BC3′ and BC3′ (this work) Figure S6. Raman spectroscopy comparison between tiled BC3′ and BC3′ (this work) Figure S7. Raman spectroscopy analysis of various BC3′ materials 11 Figure S8. B MAS NMR comparison between tiled BC3′ and BC3′ (this work) Figures S9-S10. ERDA comparison between tiled BC3′ and BC3′ (this work) Table S1. Acquisition parameters for 11B MQMAS NMR (Figure 5) Magnetic Field (T) 16.4 Temperature (K) 298 Rotor Diameter (mm) 2.5 Pulse Sequence mp3qdfs (Bruker) Number of Scans 1664 Recycle Delay (s) 0.6 Direct Dimension: 100 Spectral Width (kHz) Indirect Dimension: 125 Spinning Frequency (kHz) 20 Direct Dimension: 1024 Acquisition Length (points) Indirect Dimension: 256 Rotor Cycles for Synchronization 40 Indirect Dimension Increment (µs) 8.0 Split-t1 Increment (µs) 6.2 11B Excitation Pulse Width [π/2] (µs) 4.5 Double Frequency Sweep Length (µs) 12.5 11B Selective Pulse Width [π] (µs) 42 Table S2. Acquisition parameters for 11B MAS NMR (Figure S7) Magnetic Field (T) 16.4 Temperature (K) 298 Rotor Diameter (mm) 2.5 Pulse Sequence hahnecho (Bruker) Number of Scans 304 Recycle Delay (s) 1 Spectral Width (kHz) 100 Spinning Frequency (kHz) 20 Acquisition Length (points) 2048 11B 90° Pulse Width [π/2] (µs) 22 Table S3. -
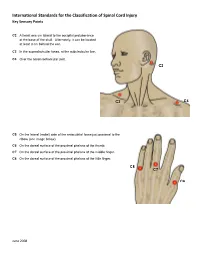
International Standards for the Classification of Spinal Cord Injury Key Sensory Points
International Standards for the Classification of Spinal Cord Injury Key Sensory Points C2 At least one cm lateral to the occipital protuberance at the base of the skull. Alternately, it can be located at least 3 cm behind the ear. C3 In the supraclavicular fossa, at the midclavicular line. C4 Over the acromioclavicular joint. C2 C3 C4 C5 On the lateral (radial) side of the antecubital fossa just proximal to the elbow (see image below). C6 On the dorsal surface of the proximal phalanx of the thumb. C7 On the dorsal surface of the proximal phalanx of the middle finger. C8 On the dorsal surface of the proximal phalanx of the little finger. C8 C7 C6 June 2008 International Standards for the Classification of Spinal Cord Injury Key Sensory Points C5 T2 T1 T1 On the medial (ulnar) side of the antecubital fossa, just proximal to the medial epicondyle of the humerus. T2 At the apex of the axilla. June 2008 International Standards for the Classification of Spinal Cord Injury Key Sensory Points T3 At the midclavicular line and the third intercostal space, found by palpating the anterior chest to locate the third C3 rib and the corresponding third intercostal space below it. T4 At the midclavicular line and the fourth intercostal space, located at the level of the nipples. T5 At the midclavicular line and the fifth intercostal space, located midway between the level of the nipples and T3 the level of the xiphisternum. T4 T6 At the midclavicular line, located at the level of the xiphisternum. T5 T7 At the midclavicular line, one quarter the distance between the level of T6 the xiphisternum and the level of the umbilicus. -

Doping Control Guide for Testing Athletes in Para Sport
DOPING CONTROL GUIDE FOR TESTING ATHLETES IN PARA SPORT JULY 2021 INTERNATIONAL PARALYMPIC COMMITTEE 2 1 INTRODUCTION This guide is intended for athletes, anti-doping organisations and sample collection personnel who are responsible for managing the sample collection process – and other organisations or individuals who have an interest in doping control in Para sport. It provides advice on how to prepare for and manage the sample collection process when testing athletes who compete in Para sport. It also provides information about the Para sport classification system (including the types of impairments) and the types of modifications that may be required to complete the sample collection process. Appendix 1 details the classification system for those sports that are included in the Paralympic programme – and the applicable disciplines that apply within the doping control setting. The International Paralympic Committee’s (IPC’s) doping control guidelines outlined, align with Annex A Modifications for Athletes with Impairments of the World Anti-Doping Agency’s International Standard for Testing and Investigations (ISTI). It is recommended that anti-doping organisations (and sample collection personnel) follow these guidelines when conducting testing in Para sport. 2 DISABILITY & IMPAIRMENT In line with the United Nations Convention on the Rights of Persons with Disabilities (CRPD), ‘disability’ is a preferred word along with the usage of the term ‘impairment’, which refers to the classification system and the ten eligible impairments that are recognised in Para sports. The IPC uses the first-person language, i.e., addressing the athlete first and then their disability. As such, the right term encouraged by the IPC is ‘athlete or person with disability’.