History and Ecology of R Pre-History the S Language
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
![R Generation [1] 25](https://docslib.b-cdn.net/cover/5865/r-generation-1-25-805865.webp)
R Generation [1] 25
IN DETAIL > y <- 25 > y R generation [1] 25 14 SIGNIFICANCE August 2018 The story of a statistical programming they shared an interest in what Ihaka calls “playing academic fun language that became a subcultural and games” with statistical computing languages. phenomenon. By Nick Thieme Each had questions about programming languages they wanted to answer. In particular, both Ihaka and Gentleman shared a common knowledge of the language called eyond the age of 5, very few people would profess “Scheme”, and both found the language useful in a variety to have a favourite letter. But if you have ever been of ways. Scheme, however, was unwieldy to type and lacked to a statistics or data science conference, you may desired functionality. Again, convenience brought good have seen more than a few grown adults wearing fortune. Each was familiar with another language, called “S”, Bbadges or stickers with the phrase “I love R!”. and S provided the kind of syntax they wanted. With no blend To these proud badge-wearers, R is much more than the of the two languages commercially available, Gentleman eighteenth letter of the modern English alphabet. The R suggested building something themselves. they love is a programming language that provides a robust Around that time, the University of Auckland needed environment for tabulating, analysing and visualising data, one a programming language to use in its undergraduate statistics powered by a community of millions of users collaborating courses as the school’s current tool had reached the end of its in ways large and small to make statistical computing more useful life. -

Data Analysis in R
Data Analysis in R Course at a Glance This course will provide an introduction to reproducible data analysis with R (see Syllabus). Instructor Gabriel Baud-Bovy ([email protected] ) Credits: 5 Synopsis This course aims at giving to the student a methodology to analyze experimental results, from how to organize data to the writing of a report. It includes: an introduction to R an introduction to reproducible research with R examples of statistical analysis with R During this course, the student will have to analyze his own data and is expected to read before each course the material that will be made available on this page. The final grade will consist in the evaluation of a report demonstrating familiarity with the concepts and methods presented in the course. As an editor, the instructor will use Notepad++ (together with NppToR) on a Windows Machines but the student might use other ones (e.g., R studio, EMACS+ESS, Lyx, TexWork). The course will use Mardown as typesetting language. For those desirous to work with Latex and/or generate pdf, you will need to install also MikeTex. Syllabus Total of 15 hours Class 1: Case study, Reproducible research Class 2: R fundamental Class 3: Exploratory data analysis and graphical methods in R Class 4: Basic statistics Class 5: To be determined There will be a final examination decided by the instructor. Prerequisites The course assumes some familiarity with programming concepts and data structures (MATLAB, C/C++, Java or any other programming language). Contact the instructor if you have never programmed anything. -

A History of R (In 15 Minutes… and Mostly in Pictures)
A history of R (in 15 minutes… and mostly in pictures) JULY 23, 2020 Andrew Zief!ler Lunch & Learn Department of Educational Psychology RMCC University of Minnesota LATIS Who am I and Some Caveats Andy Zie!ler • I teach statistics courses in the Department of Educational Psychology • I have been using R since 2005, when I couldn’t put Me (on the EPSY faculty board) SAS on my computer (it didn’t run natively on a Me Mac), and even if I could have gotten it to run, I (everywhere else) couldn’t afford it. Some caveats • Although I was alive during much of the era I will be talking about, I was not working as a statistician at that time (not even as an elementary student for some of it). • My knowledge is second-hand, from other people and sources. Statistical Computing in the 1970s Bell Labs In 1976, scientists from the Statistics Research Group were actively discussing how to design a language for statistical computing that allowed interactive access to routines in their FORTRAN library. John Chambers John Tukey Home to Statistics Research Group Rick Becker Jean Mc Rae Judy Schilling Doug Dunn Introducing…`S` An Interactive Language for Data Analysis and Graphics Chambers sketch of the interface made on May 5, 1976. The GE-635, a 36-bit system that ran at a 0.5MIPS, starting at $2M in 1964 dollars or leasing at $45K/month. ’S’ was introduced to Bell Labs in November, but at the time it did not actually have a name. The Impact of UNIX on ’S' Tom London Ken Thompson and Dennis Ritchie, creators of John Reiser the UNIX operating system at a PDP-11. -

August 12-14, Dortmund, Germany
AASC Austrian Association for Statistical Computing 2008 Program August 12-14, Dortmund, Germany useR! 2008, Dortmund, Germany 1 Contents Greetings and Miscellaneous 2 Maps 5 Social Program 8 Program 9 Tutorials 9 Schedule 10 List of Talks 14 2 useR! 2008, Dortmund, Germany Dear useRs, the following pages provide you with some useful information about useR! 2008, the R user con- ference, taking place at the Fakultät Statistik, Technische Universität Dortmund, Germany from 2008-08-12 to 2008-08-14. Pre-conference tutorials will take place on August 11. The confer- ence is organized by the Fakultät Statistik, Technische Universität Dortmund and the Austrian Association for Statistical Computing (AASC). Apart from challenging and likewise exciting scientific contributions we hope to offer you an attractive conference site and a pleasant social program. With best regards from the organizing team: Uwe Ligges (conference), Achim Zeileis (program), Claus Weihs, Gerd Kopp (local organiza- tion), Friedrich Leisch, and Torsten Hothorn Address / Contact Address: Uwe Ligges Fakultät Statistik Technische Universität Dortmund 44221 Dortmund Germany Phone: +49 231 755 4353 Fax: +49 231 755 4387 e-mail: [email protected] URL: http://www.R-Project.org/useR-2008/ Program Committee Micah Altman, Roger Bivand, Peter Dalgaard, Jan de Leeuw, Ramón Díaz-Uriarte, Spencer Graves, Leonhard Held, Torsten Hothorn, François Husson, Christian Kleiber, Friedrich Leisch, Andy Liaw, Martin Mächler, Kate Mullen, Ei-ji Nakama, Thomas Petzoldt, Martin Theus, and Heather Turner Conference Location Technische Universität Dortmund Campus Nord Mathematikgebäude / Audimax Vogelpothsweg 87 44227 Dortmund Conference Office Opening hours: Monday, August 11: 08:30–19:30 Tuesday, August 12: 08:30–18:30 Wednesday, August 13: 08:30–18:30 Thursday, August 14: 08:30–15:30 useR! 2008, Dortmund, Germany 3 Public Transport The conference site at the university campus and the city hall are best to be reached by public transport. -

Thesis Entire.Pdf (12.90Mb)
Faculty OF Science AND TECHNOLOGY Department OF Computer Science TOWARD ReprODUCIBLE Analysis AND ExplorATION OF High-ThrOUGHPUT Biological Datasets — Bjørn Fjukstad A DISSERTATION FOR THE DEGREE OF Philosophiae Doctor – 2018 This thesis document was typeset using the UiT Thesis LaTEX Template. © 2018 – http://github.com/egraff/uit-thesis “Ta aldri problemene på forskudd, for da får du dem to ganger, men ta gjerne seieren på forskudd, for hvis ikke er det altfor sjelden du får oppleve den.” –Ivar Tollefsen AbstrACT There is a rapid growth in the number of available biological datasets due to the advent of high-throughput data collection instruments combined with cheap compute infrastructure. Modern instruments enable the analysis of biological data at different levels, from small DNA sequences through larger cell structures, and up to the function of entire organs. These new datasets have brought the need to develop new software packages to enable novel insights into the underlying biological mechanisms in the development and progression of diseases such as cancer. The heterogeneity of biological datasets require researchers to tailor the explo- ration and analyses with a wide range of different tools and systems. However, despite the need for their integration, few of them provide standard inter- faces for analyses implemented using different programming languages and frameworks. In addition, because of the many tools, different input parame- ters, and references to databases, it is necessary to record these correctly. The lack of such details complicates reproducing the original results and the reuse of the analyses on new datasets. This increases the analysis time and leaves unrealized potential for scientific insights. -
Dynamic Documents with R and Knitr, by Yihui Xie 116 Tugboat, Volume 35 (2014), No
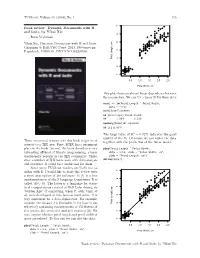
TUGboat, Volume 35 (2014), No. 1 115 ● ● ● Book review: Dynamic Documents with R ● ● ● and knitr, by Yihui Xie ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● Boris Veytsman ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● Yihui Xie, Dynamic Documents with R and knitr. ● ● ● ● ● ● ● ● ● ● ● ● ● ● Chapman & Hall/CRC Press, 2013, 190+xxvi pp. ● ● ● ● ● ● ● ● ● US$ ISBN ● Paperback, 59.95. 978-1482203530. ● ● ● ● Petal Length, cm Petal ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● 1 2 3 4 5 6 7 0.5 1.0 1.5 2.0 2.5 Petal Width, cm This plot shows an almost linear dependence between the parameters. We can try a linear fit for these data: model <- lm(Petal.Length ˜ Petal.Width, data = iris) model$coefficients ## (Intercept) Petal.Width ## 1.084 2.230 summary(model)$r.squared ## [1] 0.9271 The large value of R2 = 0.9271 indicates the good quality of the fit. Of course we can replot the data There are several reasons why this book might be of together with the prediction of the linear model: interest to a TEX user. First, LATEX has a prominent place in the book. Second, the book describes a very plot(Petal.Length ˜ Petal.Width, interesting offshoot of literate programming, a topic data = iris, xlab = "Petal Width, cm", traditionally popular in the TEX community. Third, ylab = "Petal Length, cm") abline(model) since a number of TEX users work with data analysis and statistics, R could be a useful tool for them. Since some TUGboat readers are likely not fa- ● ● ● miliar with R, I would like to start this review with ● ● a short description of the software. R [1] is a free ● ● ● ● ● ● ● ● ● ● ● implementation of the S language (sometimes R is ● ● ● ● ● ● ● ● ● ● ● ● ● ● called GNU S). -

The R Journal Volume 3/2, December 2011
The Journal Volume 3/2, December 2011 A peer-reviewed, open-access publication of the R Foundation for Statistical Computing Contents Editorial..................................................3 Contributed Research Articles Creating and Deploying an Application with (R)Excel and R...................5 glm2: Fitting Generalized Linear Models with Convergence Problems.............. 12 Implementing the Compendium Concept with Sweave and DOCSTRIP............. 16 Watch Your Spelling!........................................... 22 Ckmeans.1d.dp: Optimal k-means Clustering in One Dimension by Dynamic Programming 29 Nonparametric Goodness-of-Fit Tests for Discrete Null Distributions............... 34 Using the Google Visualisation API with R.............................. 40 GrapheR: a Multiplatform GUI for Drawing Customizable Graphs in R............. 45 rainbow: An R Package for Visualizing Functional Time Series.................. 54 Programmer’s Niche Portable C++ for R Packages...................................... 60 News and Notes R’s Participation in the Google Summer of Code 2011....................... 64 Conference Report: useR! 2011..................................... 68 Forthcoming Events: useR! 2012.................................... 70 Changes in R............................................... 72 Changes on CRAN............................................ 84 News from the Bioconductor Project.................................. 86 R Foundation News........................................... 87 2 The Journal is a peer-reviewed publication -

The R Journal, June 2012
The Journal Volume 2/1, June 2010 A peer-reviewed, open-access publication of the R Foundation for Statistical Computing Contents Editorial..................................................3 Contributed Research Articles IsoGene: An R Package for Analyzing Dose-response Studies in Microarray Experiments..5 MCMC for Generalized Linear Mixed Models with glmmBUGS ................. 13 Mapping and Measuring Country Shapes............................... 18 tmvtnorm: A Package for the Truncated Multivariate Normal Distribution........... 25 neuralnet: Training of Neural Networks............................... 30 glmperm: A Permutation of Regressor Residuals Test for Inference in Generalized Linear Models.................................................. 39 Online Reproducible Research: An Application to Multivariate Analysis of Bacterial DNA Fingerprint Data............................................ 44 Two-sided Exact Tests and Matching Confidence Intervals for Discrete Data.......... 53 Book Reviews A Beginner’s Guide to R......................................... 59 News and Notes Conference Review: The 2nd Chinese R Conference........................ 60 Introducing NppToR: R Interaction for Notepad++......................... 62 Changes in R 2.10.1–2.11.1........................................ 64 Changes on CRAN............................................ 72 News from the Bioconductor Project.................................. 85 R Foundation News........................................... 86 2 The Journal is a peer-reviewed publication of -

Opening Reproducible Research: Research Project Website and Blog Blog Post Authors
Opening Reproducible Research: research project website and blog Blog post authors: Daniel Nüst Lukas Lohoff Marc Schutzeichel Markus Konkol Matthias Hinz Rémi Rampin Vicky Steeves Opening Reproducible Research | doi:10.5281/zenodo.1485438 Blog posts Demo server update 14 Aug 2018 | By Daniel Nüst We’ve been working on demonstrating our reference-implementation during spring an managed to create a number of example workspaces. We now decided to publish these workspaces on our demo server. Screenshot 1: o2r reference implementation listing of published Executable Research Compendia . The right-hand side shows a metadata summary including original authors. The papers were originally published in Journal of Statistical Software or in a Copernicus Publications journal under open licenses. We have created an R Markdown document for each paper based on the included data and code following the ERC specification for naming core files, but only included data, an R Markdown document and a HTML display file. The publication metadata, the runtime environment description (i.e. a Dockerfile ), and the runtime image (i.e. a Docker image tarball) were all created during the ERC creation process without any human interaction (see the used R code for upload), since required metadata were included in the R Markdown document’s front matter. The documents include selected figures or in some cases the whole paper, if runtime is not extremely long. While the paper’s authors are correctly linked in the workspace metadata (see right hand side in Screenshot 1), the “o2r author” of all papers is o2r team member Daniel since he made the uploads. -

Imagej2: Imagej for the Next Generation of Scientific Image Data Curtis T
Rueden et al. BMC Bioinformatics (2017) 18:529 DOI 10.1186/s12859-017-1934-z SOFTWARE Open Access ImageJ2: ImageJ for the next generation of scientific image data Curtis T. Rueden1, Johannes Schindelin1,2, Mark C. Hiner1, Barry E. DeZonia1, Alison E. Walter1,2, Ellen T. Arena1,2 and Kevin W. Eliceiri1,2* Abstract Background: ImageJ is an image analysis program extensively used in the biological sciences and beyond. Due to its ease of use, recordable macro language, and extensible plug-in architecture, ImageJ enjoys contributions from non-programmers, amateur programmers, and professional developers alike. Enabling such a diversity of contributors has resulted in a large community that spans the biological and physical sciences. However, a rapidly growing user base, diverging plugin suites, and technical limitations have revealed a clear need for a concerted software engineering effort to support emerging imaging paradigms, to ensure the software’s ability to handle the requirements of modern science. Results: We rewrote the entire ImageJ codebase, engineering a redesigned plugin mechanism intended to facilitate extensibility at every level, with the goal of creating a more powerful tool that continues to serve the existing community while addressing a wider range of scientific requirements. This next-generation ImageJ, called “ImageJ2” in places where the distinction matters, provides a host of new functionality. It separates concerns, fully decoupling the data model from the user interface. It emphasizes integration with external applications to maximize interoperability. Its robust new plugin framework allows everything from image formats, to scripting languages, to visualization to be extended by the community. The redesigned data model supports arbitrarily large, N-dimensional datasets, which are increasingly common in modern image acquisition. -

Rtips. Revival 2012!
Rtips. Revival 2012! Paul E. Johnson <pauljohn @ ku.edu> June 8, 2012 The original Rtips started in 1999. It became difficult to update because of limitations in the software with which it was created. Now I know more about R, and have decided to wade in again. In January, 2012, I took the FaqManager HTML output and converted it to LATEX with the excellent open source program pandoc, and from there I've been editing and updating it in LYX. From here on out, the latest html version will be at http://pj.freefaculty.org/R/Rtips. html and the PDF for the same will be http://pj.freefaculty.org/R/Rtips.pdf. You are reading the New Thing! The first chore is to cut out the old useless stuff that was no good to start with, correct mistakes in translation (the quotation mark translations are particularly dangerous, but also there is trouble with ~, $, and -. Original Preface (I thought it was cute to call this \StatsRus" but the Toystore's lawyer called and, well, you know. ) If you need a tip sheet for R, here it is. This is not a substitute for R documentation, just a list of things I had trouble remembering when switching from SAS to R. Heed the words of Brian D. Ripley, \One enquiry came to me yesterday which suggested that some users of binary distributions do not know that R comes with two Guides in the doc/manual directory plus an FAQ and the help pages in book form. I hope those are distributed with all the binary distributions (they are not made nor installed by default). -

Jsr223: a Java Platform Integration for R with Programming Languages Groovy, Javascript, Jruby, Jython, and Kotlin by Floid R
CONTRIBUTED RESEARCH ARTICLES 440 jsr223: A Java Platform Integration for R with Programming Languages Groovy, JavaScript, JRuby, Jython, and Kotlin by Floid R. Gilbert and David B. Dahl Abstract The R package jsr223 is a high-level integration for five programming languages in the Java platform: Groovy, JavaScript, JRuby, Jython, and Kotlin. Each of these languages can use Java objects in their own syntax. Hence, jsr223 is also an integration for R and the Java platform. It enables developers to leverage Java solutions from within R by embedding code snippets or evaluating script files. This approach is generally easier than rJava’s low-level approach that employs the Java Native Interface. jsr223’s multi-language support is dependent on the Java Scripting API: an implementation of “JSR-223: Scripting for the Java Platform” that defines a framework to embed scripts in Java applications. The jsr223 package also features extensive data exchange capabilities and a callback interface that allows embedded scripts to access the current R session. In all, jsr223 makes solutions developed in Java or any of the jsr223-supported languages easier to use in R. Introduction About the same time Ross Ihaka and Robert Gentleman began developing R at the University of Auckland in the early 1990s, James Gosling and the so-called Green Project Team was working on a new programming language at Sun Microsystems in California. The Green Team did not set out to make a new language; rather, they were trying to move platform-independent, distributed computing into the consumer electronics marketplace. As Gosling explained, “All along, the language was a tool, not the end” (O’Connell, 1995).