Differential Regulation of E2F Trans- Activation by Cyclin/Cdk2 Complexes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Functional Compensation Among HMGN Variants Modulates the Dnase I Hypersensitive Sites at Enhancers
Downloaded from genome.cshlp.org on October 9, 2021 - Published by Cold Spring Harbor Laboratory Press Research Functional compensation among HMGN variants modulates the DNase I hypersensitive sites at enhancers Tao Deng,1,12 Z. Iris Zhu,2,12 Shaofei Zhang,1 Yuri Postnikov,1 Di Huang,2 Marion Horsch,3 Takashi Furusawa,1 Johannes Beckers,3,4,5 Jan Rozman,3,5 Martin Klingenspor,6,7 Oana Amarie,3,8 Jochen Graw,3,8 Birgit Rathkolb,3,5,9 Eckhard Wolf,9 Thure Adler,3 Dirk H. Busch,10 Valérie Gailus-Durner,3 Helmut Fuchs,3 Martin Hrabeˇ de Angelis,3,4,5 Arjan van der Velde,2,13 Lino Tessarollo,11 Ivan Ovcherenko,2 David Landsman,2 and Michael Bustin1 1Protein Section, Laboratory of Metabolism, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland 20892, USA; 2Computational Biology Branch, National Center for Biotechnology Information, National Library of Medicine, Bethesda, Maryland 20892, USA; 3German Mouse Clinic, Institute of Experimental Genetics, Helmholtz Zentrum München, German Research Center for Environmental Health, 85764 Neuherberg, Germany; 4Experimental Genetics, Center of Life and Food Sciences Weihenstephan, Technische Universität München, 85354 Freising-Weihenstephan, Germany; 5German Center for Diabetes Research (DZD), 85764 Neuherberg, Germany; 6Molecular Nutritional Medicine, Technische Universität München, 85350 Freising, Germany; 7Center for Nutrition and Food Sciences, Technische Universität München, 85350 Freising, Germany; 8Institute of Developmental Genetics (IDG), 85764 -

Cyclin-Dependent Kinases and P53 Pathways Are Activated Independently and Mediate Bax Activation in Neurons After DNA Damage
The Journal of Neuroscience, July 15, 2001, 21(14):5017–5026 Cyclin-Dependent Kinases and P53 Pathways Are Activated Independently and Mediate Bax Activation in Neurons after DNA Damage Erick J. Morris,1 Elizabeth Keramaris,2 Hardy J. Rideout,3 Ruth S. Slack,2 Nicholas J. Dyson,1 Leonidas Stefanis,3 and David S. Park2 1Massachusetts General Hospital Cancer Center, Laboratory of Molecular Oncology, Charlestown, Massachusetts 02129, 2Neuroscience Research Institute, University of Ottawa, Ottawa, Ontario K1H 8M5, Canada, and 3Columbia University, New York, New York 10032 DNA damage has been implicated as one important initiator of ization, and DNA binding that result from DNA damage are not cell death in neuropathological conditions such as stroke. Ac- affected by the inhibition of CDK activity. Conversely, no de- cordingly, it is important to understand the signaling processes crease in retinoblastoma protein (pRb) phosphorylation was that control neuronal death induced by this stimulus. Previous observed in p53-deficient neurons that were treated with camp- evidence has shown that the death of embryonic cortical neu- tothecin. However, either p53 deficiency or the inhibition of rons treated with the DNA-damaging agent camptothecin is CDK activity alone inhibited Bax translocation, cytochrome c dependent on the tumor suppressor p53 and cyclin-dependent release, and caspase-3-like activation. Taken together, our re- kinase (CDK) activity and that the inhibition of either pathway sults indicate that p53 and CDK are activated independently alone leads to enhanced and prolonged survival. We presently and then act in concert to control Bax-mediated apoptosis. show that p53 and CDKs are activated independently on par- allel pathways. -

The Involvement of Ubiquitination Machinery in Cell Cycle Regulation and Cancer Progression
International Journal of Molecular Sciences Review The Involvement of Ubiquitination Machinery in Cell Cycle Regulation and Cancer Progression Tingting Zou and Zhenghong Lin * School of Life Sciences, Chongqing University, Chongqing 401331, China; [email protected] * Correspondence: [email protected] Abstract: The cell cycle is a collection of events by which cellular components such as genetic materials and cytoplasmic components are accurately divided into two daughter cells. The cell cycle transition is primarily driven by the activation of cyclin-dependent kinases (CDKs), which activities are regulated by the ubiquitin-mediated proteolysis of key regulators such as cyclins, CDK inhibitors (CKIs), other kinases and phosphatases. Thus, the ubiquitin-proteasome system (UPS) plays a pivotal role in the regulation of the cell cycle progression via recognition, interaction, and ubiquitination or deubiquitination of key proteins. The illegitimate degradation of tumor suppressor or abnormally high accumulation of oncoproteins often results in deregulation of cell proliferation, genomic instability, and cancer occurrence. In this review, we demonstrate the diversity and complexity of the regulation of UPS machinery of the cell cycle. A profound understanding of the ubiquitination machinery will provide new insights into the regulation of the cell cycle transition, cancer treatment, and the development of anti-cancer drugs. Keywords: cell cycle regulation; CDKs; cyclins; CKIs; UPS; E3 ubiquitin ligases; Deubiquitinases (DUBs) Citation: Zou, T.; Lin, Z. The Involvement of Ubiquitination Machinery in Cell Cycle Regulation and Cancer Progression. 1. Introduction Int. J. Mol. Sci. 2021, 22, 5754. https://doi.org/10.3390/ijms22115754 The cell cycle is a ubiquitous, complex, and highly regulated process that is involved in the sequential events during which a cell duplicates its genetic materials, grows, and di- Academic Editors: Kwang-Hyun Bae vides into two daughter cells. -

HMGB1 in Health and Disease R
Donald and Barbara Zucker School of Medicine Journal Articles Academic Works 2014 HMGB1 in health and disease R. Kang R. C. Chen Q. H. Zhang W. Hou S. Wu See next page for additional authors Follow this and additional works at: https://academicworks.medicine.hofstra.edu/articles Part of the Emergency Medicine Commons Recommended Citation Kang R, Chen R, Zhang Q, Hou W, Wu S, Fan X, Yan Z, Sun X, Wang H, Tang D, . HMGB1 in health and disease. 2014 Jan 01; 40():Article 533 [ p.]. Available from: https://academicworks.medicine.hofstra.edu/articles/533. Free full text article. This Article is brought to you for free and open access by Donald and Barbara Zucker School of Medicine Academic Works. It has been accepted for inclusion in Journal Articles by an authorized administrator of Donald and Barbara Zucker School of Medicine Academic Works. Authors R. Kang, R. C. Chen, Q. H. Zhang, W. Hou, S. Wu, X. G. Fan, Z. W. Yan, X. F. Sun, H. C. Wang, D. L. Tang, and +8 additional authors This article is available at Donald and Barbara Zucker School of Medicine Academic Works: https://academicworks.medicine.hofstra.edu/articles/533 NIH Public Access Author Manuscript Mol Aspects Med. Author manuscript; available in PMC 2015 December 01. NIH-PA Author ManuscriptPublished NIH-PA Author Manuscript in final edited NIH-PA Author Manuscript form as: Mol Aspects Med. 2014 December ; 0: 1–116. doi:10.1016/j.mam.2014.05.001. HMGB1 in Health and Disease Rui Kang1,*, Ruochan Chen1, Qiuhong Zhang1, Wen Hou1, Sha Wu1, Lizhi Cao2, Jin Huang3, Yan Yu2, Xue-gong Fan4, Zhengwen Yan1,5, Xiaofang Sun6, Haichao Wang7, Qingde Wang1, Allan Tsung1, Timothy R. -

A Haploid Genetic Screen Identifies the G1/S Regulatory Machinery As a Determinant of Wee1 Inhibitor Sensitivity
A haploid genetic screen identifies the G1/S regulatory machinery as a determinant of Wee1 inhibitor sensitivity Anne Margriet Heijinka, Vincent A. Blomenb, Xavier Bisteauc, Fabian Degenera, Felipe Yu Matsushitaa, Philipp Kaldisc,d, Floris Foijere, and Marcel A. T. M. van Vugta,1 aDepartment of Medical Oncology, University Medical Center Groningen, University of Groningen, 9723 GZ Groningen, The Netherlands; bDivision of Biochemistry, The Netherlands Cancer Institute, 1066 CX Amsterdam, The Netherlands; cInstitute of Molecular and Cell Biology, Agency for Science, Technology and Research, Proteos#3-09, Singapore 138673, Republic of Singapore; dDepartment of Biochemistry, National University of Singapore, Singapore 117597, Republic of Singapore; and eEuropean Research Institute for the Biology of Ageing, University of Groningen, University Medical Center Groningen, 9713 AV Groningen, The Netherlands Edited by Stephen J. Elledge, Harvard Medical School, Boston, MA, and approved October 21, 2015 (received for review March 17, 2015) The Wee1 cell cycle checkpoint kinase prevents premature mitotic Wee1 kinase at tyrosine (Tyr)-15 to prevent unscheduled Cdk1 entry by inhibiting cyclin-dependent kinases. Chemical inhibitors activity (5, 6). Conversely, timely activation of Cdk1 depends on of Wee1 are currently being tested clinically as targeted anticancer Tyr-15 dephosphorylation by one of the Cdc25 phosphatases drugs. Wee1 inhibition is thought to be preferentially cytotoxic in (7–10). When DNA is damaged, the downstream DNA damage p53-defective cancer cells. However, TP53 mutant cancers do not response (DDR) kinases Chk1 and Chk2 inhibit Cdc25 phos- respond consistently to Wee1 inhibitor treatment, indicating the phatases through direct phosphorylation, which blocks Cdk1 existence of genetic determinants of Wee1 inhibitor sensitivity other activation (11–13). -
Supplemental Data.Pdf
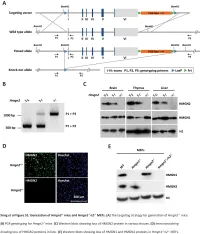
Table S1. Summary of sequencing results A. DNase‐seq data % Align % Mismatch % >=Q30 bases Sample ID # Reads % unique reads (PF) Rate (PF) (PF) WT_1 84,301,522 86.76 0.52 93.34 96.30 WT_2 98,744,222 84.97 0.51 93.75 89.94 Hmgn1‐/‐_1 79,620,656 83.94 0.86 90.58 88.65 Hmgn1‐/‐_2 62,673,782 84.13 0.87 91.30 89.18 Hmgn2‐/‐_1 87,734,440 83.49 0.71 91.81 90.00 Hmgn2‐/‐_2 82,498,808 83.25 0.69 92.73 90.66 Hmgn1‐/‐n2‐/‐_1 71,739,638 68.51 2.31 81.11 89.22 Hmgn1‐/‐n2‐/‐_2 74,113,682 68.19 2.37 81.16 86.57 B. ChIP‐seq data Histone % Align % Mismatch % >=Q30 % unique Genotypes # Reads marks (PF) Rate (PF) bases (PF) reads H3K4me1 100670054 92.99 0.28 91.21 87.29 H3K4me3 67064272 91.97 0.35 89.11 27.15 WT H3K27ac 90,340,242 93.57 0.28 95.02 89.80 input 111,292,572 78.24 0.55 96.07 86.99 H3K4me1 84598176 92.34 0.33 91.2 81.69 H3K4me3 90032064 92.19 0.44 88.76 15.81 Hmgn1‐/‐n2‐/‐ H3K27ac 86,260,526 93.40 0.29 94.94 87.49 input 78,142,334 78.47 0.56 95.82 81.11 C. MNase‐seq data % Mismatch % >=Q30 bases % unique Sample ID # Reads % Align (PF) Rate (PF) (PF) reads WT_1_Extensive 45,232,694 55.23 1.49 90.22 81.73 WT_1_Limited 105,460,950 58.03 1.39 90.81 79.62 WT_2_Extensive 40,785,338 67.34 1.06 89.76 89.60 WT_2_Limited 105,738,078 68.34 1.05 90.29 85.96 Hmgn1‐/‐n2‐/‐_1_Extensive 117,927,050 55.74 1.49 89.50 78.01 Hmgn1‐/‐n2‐/‐_1_Limited 61,846,742 63.76 1.22 90.57 84.55 Hmgn1‐/‐n2‐/‐_2_Extensive 137,673,830 60.04 1.30 89.28 78.99 Hmgn1‐/‐n2‐/‐_2_Limited 45,696,614 62.70 1.21 90.71 85.52 D. -

Cyclin a Triggers Mitosis Either Via Greatwall Or Cyclin B
bioRxiv preprint doi: https://doi.org/10.1101/501684; this version posted December 20, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Cyclin A triggers Mitosis either via Greatwall or Cyclin B Nadia Hégarat(1)*, Adrijana Crncec(1)*, Maria F. Suarez Peredoa Rodri- guez(1), Fabio Echegaray Iturra(1), Yan Gu(1), Paul F. Lang(2), Alexis R. Barr(3), Chris Bakal(4), Masato T. Kanemaki(5), Angus I. Lamond(6), Bela Novak(2), Tony Ly(7)•• and Helfrid Hochegger(1)•• (1) Genome Damage and Stability Centre, School of Life Sciences, University of Sussex, Brighton BN19RQ, UK (2) Department of Biochemistry, University of Oxford, South Park Road, Oxford OX13QU, UK (3) MRC London Institute of Medical Science, Imperial College, London W12 0NN, UK (4) The Institute of Cancer Research, London SW3 6JB, UK (5) National Institute of Genetics, Research Organization of Information and Sys- tems (ROIS), and Department of Genetics, SOKENDAI (The Graduate University of Advanced Studies), Yata 1111, Mishima, Shizuoka 411-8540, Japan. (6) Centre for Gene Regulation and Expression, School of Life Sciences, University of Dundee, Dundee DD1 5EH, UK (7) Wellcome Trust Centre for Cell Biology, University of Edinburgh, Edinburgh EH9 3BF, UK * Equal contribution ** Correspondence: Tony Ly: [email protected]; Helfrid Hochegger: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/501684; this version posted December 20, 2018. -

Cytometry of Cyclin Proteins
Reprinted with permission of Cytometry Part A, John Wiley and Sons, Inc. Cytometry of Cyclin Proteins Zbigniew Darzynkiewicz, Jianping Gong, Gloria Juan, Barbara Ardelt, and Frank Traganos The Cancer Research Institute, New York Medical College, Valhalla, New York Received for publication January 22, 1996; accepted March 11, 1996 Cyclins are key components of the cell cycle pro- gests that the partner kinase CDK4 (which upon ac- gression machinery. They activate their partner cy- tivation by D-type cyclins phosphorylates pRB com- clin-dependent kinases (CDKs) and possibly target mitting the cell to enter S) is perpetually active them to respective substrate proteins within the throughout the cell cycle in these tumor lines. Ex- cell. CDK-mediated phosphorylation of specsc sets pression of cyclin D also may serve to discriminate of proteins drives the cell through particular phases Go vs. GI cells and, as an activation marker, to iden- or checkpoints of the cell cycle. During unper- tify the mitogenically stimulated cells entering the turbed growth of normal cells, the timing of expres- cell cycle. Differences in cyclin expression make it sion of several cyclins is discontinuous, occurring possible to discrirmna* te between cells having the at discrete and well-defined periods of the cell cy- same DNA content but residing at different phases cle. Immunocytochemical detection of cyclins in such as in G2vs. M or G,/M of a lower DNA ploidy vs. relation to cell cycle position (DNA content) by GI cells of a higher ploidy. The expression of cyclins multiparameter flow cytometry has provided a new D, E, A and B1 provides new cell cycle landmarks approach to cell cycle studies. -

Binding RXFP1 in Brain Tumors
Functional role of C1Q-TNF related peptide 8 (CTRP8)- binding RXFP1 in brain tumors By THATCHAWAN THANASUPAWAT A Thesis submitted to the Faculty of Graduate Studies of The University of Manitoba in partial fulfillment of the requirements of the degree of DOCTOR OF PHILOSOPHY Department of Human Anatomy and Cell Science University of Manitoba Winnipeg, Manitoba Canada Copyright © 2017 by Thatchawan Thanasupawat TABLE OF CONTENTS ABSTRACT ....................................................................................................................... v ACKNOWLEDGEMENTS ........................................................................................... vii LIST OF TABLES ........................................................................................................... ix LIST OF FIGURES .......................................................................................................... x LIST OF ABBREVIATIONS ........................................................................................ xii LIST OF COPYRIGHTED MATERIALS ................................................................. xix CHAPTER 1: INTRODUCTION .................................................................................... 1 1.1 Relaxin family peptide receptors (RXFPs) ............................................................... 1 1.2 Relaxin family peptide receptor 1 (RXFP1) ............................................................. 1 1.2.1 Structural features of RXFP1 ............................................................................ -

Identification of Proteins Involved in the Maintenance of Genome Stability
Identification of Proteins Involved in the Maintenance of Genome Stability by Edith Hang Yu Cheng A thesis submitted in conformity with the requirements for the degree of Doctor of Philosophy Department of Biochemistry University of Toronto ©Copyright by Edith Cheng2015 Identification of Proteins Involved in the Maintenance of Genome Stability Edith Cheng Doctor of Philosophy Department of Biochemistry University of Toronto 2015 Abstract Aberrant changes to the genome structure underlie numerous human diseases such as cancers. The functional characterization ofgenesand proteins that maintain chromosome stability will be important in understanding disease etiology and developing therapeutics. I took a multi-faceted approach to identify and characterize genes involved in the maintenance of genome stability. As biological pathways involved in genome maintenance are highly conserved in evolution, results from model organisms can greatly facilitate functional discovery in humans. In S. cerevisiae, I identified 47 essential gene depletions with elevated levels of spontaneous DNA damage foci and 92 depletions that caused elevated levels of chromosome rearrangements. Of these, a core subset of 15 DNA replication genes demonstrated both phenotypes when depleted. Analysis of rearrangement breakpoints revealed enrichment at yeast fragile sites, Ty retrotransposons, early origins of replication and replication termination sites. Together, thishighlighted the integral role of DNA replicationin genome maintenance. In light of my findings in S. cerevisiae, I identified a list of 153 human proteins that interact with the nascentDNA at replication forks, using a DNA pull down strategy (iPOND) in human cell lines. As a complementary approach for identifying human proteins involved in genome ii maintenance, I usedthe BioID techniqueto discernin vivo proteins proximal to the human BLM- TOP3A-RMI1-RMI2 genome stability complex, which has an emerging role in DNA replication progression. -

The Hmgn Family of Chromatin Binding Proteins Regulate Stem Cell Pluripotency and Neuronal Differentiation
The Hmgn family of chromatin binding proteins regulate stem cell pluripotency and neuronal differentiation Gokula Mohan, Ohoud Rehbini, Tomoko Iwata, Katherine West Institute of Cancer Sciences, University of Glasgow Hmgn proteins Hmgn proteins are enriched across Hmgn2 is expressed in neural progenitor cells the Oct4 and Nanog gene loci during embryogenesis E11.5 E13.5 E15.5 Bind as dimers to nucleosomes svz Hmgn1 Hmgn2 svz 3 Regulate the deposition of histone 3 3 2.5 modifications 2.5 2.5 2 2 2 1.5 Compete with linker histones 1.5 1.5 1 enrichment 1 1 Interact with several transcription 0.5 0.5 0.5 factors. 0 0 0 svz: ventricular/ -1781 -399 410 2070 3342 -1081 -219 929 1740 2636 Actin Msat c subventricular zone Kato H et al. PNAS 2011;108:12283-12288 Oct4 Nanog H3 dg dg: dentate gyrus (in hippocampus) control cells NLS Nucleosome-binding domain NLS Regulatory domain 2 2 2 very highly conserved Competes with H1 c: cerebellum 1.5 1.5 1.5 100 aa 1 1 1 P14 Inhibits phosphorylation Increases acetylation enrichment 0.5 0.5 0.5 of H3S10 of H3K14 0 0 0 -1781 -399 410 2070 3342 -1081 -219 929 1740 2636 Actin Msat Retinoic acid-induced differentiation of EC P Oct4 Nanog cells down the neuronal lineage. M A A P Reprogramming Differentiation Hmgn1 and Hmgn2 binding profiles in control cells. Enrichment at each PCR set was normalised to Histone H3 N-terminal tail ARTKQTARKSTGGKAPRKQLATKAARKSA... stage stage input and then normalised to average H3 enrichment across all primer sets to control for differences M between chromatin preps. -

HMGA2-Mediated Epigenetic Regulation of Gata6 Controls Epithelial Canonical WNT Signaling During Lung Development and Homeostasis
HMGA2-mediated epigenetic regulation of Gata6 controls epithelial canonical WNT signaling during lung development and homeostasis INAUGURALDISSERTATION zur Erlangung des Doktorgrades der Naturwissenschaften - Doctor rerum naturalium - (Dr. rer. nat.) eingereicht am Fachbereich Biologie und Chemie der Justus-Liebig-Universität Giessen vorgelegt von Indrabahadur Singh Bad Nauheim, 2014 Die vorliegende Arbeit wurde am Max-Planck-Institut für Herz- und Lungenforschung in der Abteilung "Epigenetik des Lungenkrebs" unter der Leitung von Herrn Dr. Guillermo Barreto angefertigt. Erstgutachter: Prof. Dr. Dr. Thomas Braun Abteilung Entwicklung und Umbau des Herzens Max-Planck-Institut für Herz- und Lungenforschung Ludwigstraße 43, 61231 Bad Nauheim Zweitgutachter: Prof. Dr. Rainer Renkawitz Institut für Genetik Justus-Liebig-Universität Giessen Heinrich-Buff-Ring 58, 35392 Giessen Disputation am 24-09-2014 DEDICATED TO MY PARENTS & GRANDPARENTS Whose perpetual affection and blessing always Inspired me for higher ambition in life TABLE OF CONTENTS Table of Contents Table of Contents ....................................................................................................................... VIII List of Figures and Tables............................................................................................................. XI ZUSAMMENFASSUNG ......................................................................................................... XIV ABSTRACT ..................................................................................................................................