Creating a Web-Based 3D Interactive Resource to Teach the Anatomy of the Human Hippocampus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A. HM, B. Spa@Al Naviga@On 2. Hippocampus
Outline 1. What does Hippocampus do? A. HM, B. Spaal navigaon 2. Hippocampus anatomy and connec;vity CA1, CA3 diagram etc. Connec;vity to other regions. 3. Place fields in Hippocampus – show examples + the O'Keefe effect 4. Plas;city of place fields + NMDA blocker + Morris water maze 5. Replay of sequences 6. Entorhinal grid cells What does the Hippocampus do? 1. Memory 2. Spaal representaon Henry Gustav Molaison (February 26, 1926 – December 2, 2008), beger known as HM “AYer operaon this young man could no longer recognize the hospital staff nor find his way to the bathroom, and he seemed to recall nothing of the day-to-day events of his hospital life. There was also a par;al retrograde amnesia, inasmuch as he did not remember the death of a favorite uncle three years previously, nor anything of the period in the hospital, yet could recall some trivial events that had occurred just before his admission to the hospital. His early memories were apparently vivid and intact. This paent’s memory defect has persisted without improvement to the present ;me, and numerous illustraons of its severity could be given. Ten months ago the family moved from their old house to a new one a few blocks away on the same street; he s;ll has not learned the new address, though remembering the old one perfectly, nor can he be trusted to find his way home alone.” “Moreover, he does not know where objects in con;nual use are kept; for example, his mother s;ll has to tell him where to find the lawn mower, even though he may have been using it only the day before. -

Retrohippocampal Cortical Neurons During Hippocampal Sharp Waves in the Behaving Rat
The Journal of Neuroscience, October 1994, 74(10): 6160-6170 Selective Activation of Deep Layer (V-VI) Retrohippocampal Cortical Neurons during Hippocampal Sharp Waves in the Behaving Rat J. J. Chrobak and G. Buzsaki Center for Molecular and Behavioral Neuroscience, Rutgers, The State University of New Jersey, Newark, New Jersey 07102 The coordinated activity of hippocampal neurons is reflected feet on their postsynaptic neocortical targets and may rep- by macroscopic patterns, theta and sharp waves (SPW), ev- resent a physiological mechanism for memory trace transfer ident in extracellular field recordings. The importance of these from the hippocampus to the neocortex. patterns is underscored by the ordered relation of specific [Key words: hippocampus, entorhinal cortex, theta, sharp neuronal populations to each pattern as well as the relation waves, oscillations, memory, temporal lobe epilepsy, Ab- of each pattern to distinct behavioral states. During awake heimer’s disease] immobility, consummatory behavior, and slow wave sleep, CA3 and CA1 neurons participate in organized population Retrohippocampal structures [entorhinal cortex (EC), parasub- bursts during SPW. In contrast, during theta-associated ex- iculum, presubiculum, and subiculum] process and transmit ploratory activity, the majority of principle cells are silent. information between the neocortex and the hippocampus. The Considerably less is known about the discharge properties electrophysiology of thesestructures has received scant attention of retrohippocampal neurons during theta, and particularly despite their importance as a substrate for memory (Amaral, during SPW. These retrohippocampal neurons (entorhinal 1987; Zola-Morgan et al., 1989; Squire, 1992)and asfocal point cortical, parasubicular, presubicular, and subicular) process for the pathophysiology of dementia (Hyman et al., 1984; Van and transmit information between the neocortex and the hip- Hoesenet al., 1991) and temporal lobe epilepsy (Rutecki et al., pocampus. -

Traumatic Early Life Stress in the Developing Hippocampus: a Meta-Analysis of MRI Studies
Walden University ScholarWorks Walden Dissertations and Doctoral Studies Walden Dissertations and Doctoral Studies Collection 2020 Traumatic Early Life Stress in the Developing Hippocampus: A Meta-Analysis of MRI Studies Sharon Johnson Walden University Follow this and additional works at: https://scholarworks.waldenu.edu/dissertations Part of the Psychology Commons This Dissertation is brought to you for free and open access by the Walden Dissertations and Doctoral Studies Collection at ScholarWorks. It has been accepted for inclusion in Walden Dissertations and Doctoral Studies by an authorized administrator of ScholarWorks. For more information, please contact [email protected]. Walden University College of Social and Behavioral Sciences This is to certify that the doctoral dissertation by Sharon Lee Johnson has been found to be complete and satisfactory in all respects, and that any and all revisions required by the review committee have been made. Review Committee Dr. Scott Wowra, Committee Chairperson, Psychology Faculty Dr. Patricia Costello, Committee Member, Psychology Faculty Dr. Kimberley Cox, University Reviewer, Psychology Faculty Chief Academic Officer and Provost Sue Subocz, Ph.D. Walden University 2020 Abstract Traumatic Early Life Stress in the Developing Hippocampus: A Meta-Analysis of MRI Studies by Sharon Lee Johnson MPhil, Walden University, 2019 M.Ed., Youngstown State University, 1994 BA, Kent State University, 1979 Dissertation Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Health Psychology Walden University August 2020 Abstract Advancements in neuroimaging techniques afford researchers the opportunity to examine the actual brains of living persons, which exponentially contributes to new insights regarding brain and behavior phenomena. However, empirical studies investigating stress and the hippocampus attend primarily to adult populations - less on children and adolescents. -

Diseño Y Desarrollo De Un Sitio Web E-Commerce Para La Distribución Y Comercialización De Piezas Gráficas Tridimensionales Producidas En Blender®
Diseño y desarrollo de un sitio web e-commerce para la distribución y comercialización de piezas gráficas tridimensionales producidas en Blender®. Memoria de Proyecto Final de Grado/Máster Máster Universitario en Aplicaciones multimedia Informática, multimedia y telecomunicación Autor: Juan Pablo Ruiz Yépez Consultor: Mikel Zorrilla Berasategui Profesor: Laura Porta Simó 7 junio 2019 Diseño y desarrollo de un sitio web e-commerce para la distribución y comercialización de piezas gráficas tridimensionales producidas en Blender®, Máster en Aplicaciones Multimedia, Juan Pablo Ruiz Yépez Créditos/Copyright Esta obra está sujeta a una licencia de Reconocimiento-NoComercial-SinObraDerivada 3.0 España de CreativeCommons 2 / 53 Diseño y desarrollo de un sitio web e-commerce para la distribución y comercialización de piezas gráficas tridimensionales producidas en Blender®, Máster en Aplicaciones Multimedia, Juan Pablo Ruiz Yépez FICHA DEL TRABAJO FINAL Diseño y desarrollo de un sitio web e-commerce para Título del trabajo: la distribución y comercialización de piezas gráficas tridimensionales producidas en Blender®, Nombre del autor: Juan Pablo Ruiz Yépez Nombre del consultor/a: Mikel Zorrilla Berasategui Nombre del PRA: Laura Porta Simó Fecha de entrega (mm/aaaa): 06/2019 Titulación: Máster en Aplicaciones Multimedia Área del Trabajo Final: Trabajo Fin de Máster Idioma del trabajo: Español Palabras clave Modelos 3d, e-commerce, blender Resumen del Trabajo: Este trabajo tiene como fin la creación de un sitio web adaptivo basado en la plataforma e-commerce para la distribución de objetos tridimensionales creados con la aplicación Blender®. El sitio ha sido desarrollado con la tecnología html5 y el apoyo de la plataforma Wordpress, para finalmente ser alojado en un hosting compartido. -

Webar Development Tools: an Overview
WebAR development tools: An overview Dmytro S. Shepilieva, Yevhenii O. Modlod, Yuliia V. Yechkalob, Viktoriia V. Tkachukb, Mykhailo M. Mintiia, Iryna S. Mintiia,c, Oksana M. Markovab, Tetiana V. Selivanovaa, Olena M. Drashkoa, Olga O. Kalinichenkoa, Tetiana A. Vakaliukb,e, Viacheslav V. Osadchyif and Serhiy O. Semerikova,b,c aKryvyi Rih State Pedagogical University, 54 Gagarin Ave., Kryvyi Rih, 50086, Ukraine bKryvyi Rih National University, 11 Vitalii Matusevych Str., Kryvyi Rih, 50027, Ukraine cInstitute of Information Technologies and Learning Tools of the NAES of Ukraine, 9 M. Berlynskoho Str., Kyiv, 04060, Ukraine dState University of Economics and Technology, 5 Stephana Tilhy Str., Kryvyi Rih, 50006, Ukraine eZhytomyr Polytechnic State University, 103 Chudnivska Str., Zhytomyr, 10005, Ukraine fBogdan Khmelnitsky Melitopol State Pedagogical University, 20 Hetmanska Str., Melitopol, 72300, Ukraine Abstract Web augmented reality (WebAR) development tools aimed at improving the visual aspects of learning are far from being visual and available themselves. This causing problems of selecting and testing WebAR development tools for CS undergraduates mastering in web-design basics. The research is aimed at conducting comparative analysis of WebAR tools to select those appropriated for beginners. Keywords augmented reality software tools, WebAR, A-Frame, AR.js, Three.js, JSARToolKit CS&SE@SW 2020: 3rd Workshop for Young Scientists in Computer Science & Software Engineering, November 27, 2020, Kryvyi Rih, Ukraine " [email protected] (D.S. Shepiliev); [email protected] (Y.O. Modlo); [email protected] (Y.V. Yechkalo); [email protected] (V.V. Tkachuk); [email protected] (M.M. Mintii); [email protected] (I.S. Mintii); [email protected] (O.M. -
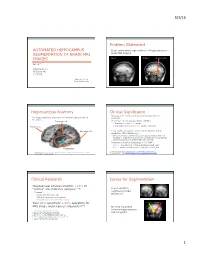
5/3/16 1 Automated Hippocampus Segmentation of Brain
5/3/16 Problem Statement AUTOMATED HIPPOCAMPUS • Goal: automated segmentation of hippocampus in SEGMENTATION OF BRAIN MRI brain MRI images Input Output hippocampus IMAGES Group 7 Yingchuan Hu Chunyan Wu Li Zhong May 1st, 2014 Cornell University Hippocampus Anatomy Clinical Significance • Epilepsy is the most common serious brain disorder The hippocampus is located in the medial temporal lobe of worldwide. the brain. 1 Temporal lobe • Prevalence of epilepsy worldwide (WHO ) • 7 sufferers in every 1,000 people • 3 new sufferers in every 10,000 people each year Frontal lobe Occipital lobe • People with epilepsy are at increased risks for status epilepticus (life-threatening) • One continuous, unremitting seizure lasting longer than five minutes or recurrent seizures without regaining consciousness between seizures for greater than five minutes. • Prevalence of status epilepticus in US (NIH2) • 195,000 new patients of status epilepticus each year • 42,000 deaths caused by status epilepticus each year 1. World Health Organization http://www.who.int/mental_health/neurology/epilepsy/en/ *Modified from a scan of a plate of “Posterior and inferior cornua of left lateral ventricle exposed 2. National institute of Health http://www.ninds.nih.gov/disorders/epilepsy/detail_epilepsy.htm from the side” in Gary’s Anatomy Clinical Research Issues for Segmentation • Hippocampal volume reduction >10% of “normal” size indicates epilepsy.[1-4] • Low contrast to neighboring brain • “normal”: structures • People with the same age • Bilateral hippocampus comparison • Personal changes in more than 1 year • Over 90% sensitivity + 98% specificity for amgydala hippocampus [5-7] MRI image measurement diagnosis. • No clear boundary between hippocampus 1. Cook MJ, et al. -

Annual Report 2018
ANNUAL REPORT 2017-2018 Table of Contents 2 Director’s Letter 3 History and Mission 4 Faculty 8 Faculty News 12 Faculty Research Programs 22 Scientific Advancements 34 Collaborations 46 Academic and Community Activities 51 Faculty Publications FY 2018 59 Teaching 62 Postdoctoral Trainees 64 Visiting Scientists 65 Graduate Students 69 Grants and Contracts 77 Events 77 Zilkha Seminar Series 80 8th Annual Zach Hall Lecture 82 Los Angeles Brain Bee 84 Soprano and Musical Ambassador Renée Fleming visits Zilkha 85 Music to Remember – LA Opera/Alzheimer’s Greater Los Angeles 86 Searching for Solutions: 5th Annual AD Symposia Held at Zilkha Institute 87 Administration and Operating Budget 88 Development 1 Director’s Letter Dear Friends, The year 2018 began with a strong start when the department of Physiology & Neuroscience was ranked number 3 in the nation according to the Blue Ridge Institute. I am proud of the work we have done to achieve this great honor and in awe of our faculty who have made this accomplishment possible, by receiving a large number of federal awards for their research. Of course, as much as we love to have well-funded research programs, it is the science that really matters, and as you will see across the following pages, we have made great strides in so many different areas, from neural circuits and genomics, to imaging and Alzheimer’s. I am proud of the work we are doing and happy to be recognized for it. In addition to awards received, we welcomed a number of visitors and speakers who came to the Zilkha Neurogenetic Institute, some to share their cutting-edge research, others to learn of the advances we have made. -

Mapping the Mechanobiome: Novel Mathematically-Derived 3D Visualization of the Cellular Mechanoresponsive System for Interactive Publication
Mapping the Mechanobiome: Novel mathematically-derived 3D visualization of the cellular mechanoresponsive system for interactive publication by Cecilia C. Johnson A thesis submitted to Johns Hopkins University in conformity with the requirements for the degree of Master of Arts. Baltimore, Maryland March, 2019 © 2019 Cecilia Johnson All Rights Reserved Abstract Mechanical forces, ubiquitous in biological settings, are major determinants of cell fate; they should not be considered a detail applicable to specialized circumstances but rather a vital component of cell biology. To sense, respond, and generate both intracellular and extracellular mechanical forces, cells contain a highly integrated and dynamic network of macromolecules throughout the cell. The Robinson Lab at the Johns Hopkins School of Medicine developed the term “mechanobiome,” to describe and categorize that network of macromolecules. At the interface of cell biology, physics, and engineering, the concept of the mechanobiome provides researchers a systems-level understanding of the extensive contributions of physical force and mechanical cell properties on cell morphology, differentiation, physiology, and disease. Although numerous diseases, including cancer, cardiovascular disease, and chronic obstructive pulmonary disease, develop from abnormal cell mechanics, the mechanobiome is rarely explored as a novel source of therapeutic targets. Increased understanding of the mechanobiome will enhance understanding of normal biological machinery and ultimately lead to new pathways for targeting disease. To address the lack of comprehensive, accurate visualizations of the mechanobiome, two novel theoretical 3D models of the mechanobiome were developed: one at the cellular level and one at the nanoscale level. By integrating published data on components of the mechanobiome, such as crystal structures, macromolecule concentrations, and polymer dissociation constants, a proportionately accurate visualization of the cell’s mechanical system was produced. -

Ostvarivanje 3D Grafike U Internet Preglednicima Pomoću Biblioteke Three.Js
Ostvarivanje 3D grafike u Internet preglednicima pomoću biblioteke Three.js Ivanušec, Sandi Master's thesis / Diplomski rad 2021 Degree Grantor / Ustanova koja je dodijelila akademski / stručni stupanj: University of Pula / Sveučilište Jurja Dobrile u Puli Permanent link / Trajna poveznica: https://urn.nsk.hr/urn:nbn:hr:137:571280 Rights / Prava: In copyright Download date / Datum preuzimanja: 2021-10-06 Repository / Repozitorij: Digital Repository Juraj Dobrila University of Pula Sveučilište Jurja Dobrile u Puli Fakultet informatike u Puli SANDI IVANUŠEC OSTVARIVANJE 3D GRAFIKE U INTERNET PREGLEDNICIMA POMOĆU BIBLIOTEKE THREE.JS Diplomski rad Pula, lipanj, 2021. Sveučilište Jurja Dobrile u Puli Fakultet informatike u Puli SANDI IVANUŠEC OSTVARIVANJE 3D GRAFIKE U INTERNET PREGLEDNICIMA POMOĆU BIBLIOTEKE THREE.JS Diplomski rad JMBAG: 0303069339, redoviti student Studijski smjer: Informatika Predmet: Izrada informatičkih projekata Mentor: doc. dr. sc. Nikola Tanković Pula, lipanj, 2021. IZJAVA O AKADEMSKOJ ČESTITOSTI Ja, dolje potpisani _________________________, kandidat za magistra ______________________________________________ovime izjavljujem da je ovaj Diplomski rad rezultat isključivo mojega vlastitog rada, da se temelji na mojim istraživanjima te da se oslanja na objavljenu literaturu kao što to pokazuju korištene bilješke i bibliografija. Izjavljujem da niti jedan dio Diplomskog rada nije napisan na nedozvoljen način, odnosno da je prepisan iz kojega necitiranog rada, te da ikoji dio rada krši bilo čija autorska prava. Izjavljujem, -

Hippocampus and Shape Analysis Claire Cury
Hippocampus and shape analysis Claire Cury To cite this version: Claire Cury. Hippocampus and shape analysis. TIG Symposium for Epilepsy, Mar 2017, London, United Kingdom. hal-02080594 HAL Id: hal-02080594 https://hal.inria.fr/hal-02080594 Submitted on 26 Mar 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Hippocampus and shape analysis TIG symposium for epilepsy Claire Cury Introduction • Position: Position of the object in its environment. • Shape: 3D edge of an object. Ridge transformation invariant. • Statistical shape analysis of anatomical structures – Modelisation of normal and pathological variability. – Prediction of clinical and biological parameters. Hippocampus Anatomy Images: Duvernoy et al, 2005 Incomplete Hippocampal Inversion (IHI) Mainly described in epileptic patients (Barsi et al. 2000; Bajic et al. 2009;…) ~ 50% In healthy population (Bernasconi et al. 2005; Bajic et al. 2008; Gamss et al. 2009) ~ 20% Criteria ill-defined Mix of healthy and controls (non epileptics) (Gamss et al. 2009) Not enough subjects (Bajic et al. 2008) IHI : Criteria • C1: roundness and verticality • C2: collateral sulci • C3: position • C4: subiculum • C5: T4 sulci • C0, IHI global appraisal: – 0 : normal aspect – 1 : Partial IHI – 2 : Total IHI Cury et al. -

Digital Photogrammetry, Tls Survey and 3D Modelling for Vr and Ar Applications in Ch
The International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences, Volume XLIII-B2-2020, 2020 XXIV ISPRS Congress (2020 edition) DIGITAL PHOTOGRAMMETRY, TLS SURVEY AND 3D MODELLING FOR VR AND AR APPLICATIONS IN CH. A. Scianna 1, *, G. F. Gaglio 2 , M. La Guardia 3 1ICAR-CNR (High Performance Computing and Networking Institute - National Research Council of Italy) at GISLab c/o D’Arch, Polytechnic School of University of Palermo, Viale delle Scienze, Edificio 8, 90128 Palermo, Italy [email protected] 2 ICAR-CNR (High Performance Computing and Networking Institute - National Research Council of Italy), at GISLab, Via Ugo La Malfa 153, Edificio 8, 90146 Palermo, Italy [email protected] 3 Department of Engineering, Polytechnic School of University of Palermo, Viale delle Scienze, Edificio 10, 90128 Palermo, Italy, [email protected] Commission II KEY WORDS: AR, VR, TLS, UAV, Photogrammetry, 3D modelling. ABSTRACT: The world of valorization of Cultural Heritage is even more focused on the virtual representation and reconstructions of digital 3D models of monuments and archaeological sites. In this scenario the quality and the performances offered by the virtual reality (VR) and augmented reality (AR) navigation take primary importance, improving the accessibility of cultural sites where the real access is not allowed for natural conditions or human possibilities. The creation of a virtual environment useful for these purposes requires a specific workflow to follow, combining different strategies in the fields of survey, 3D modelling and virtual navigation. In this work a specific case of study has been analyzed as a practical example, the church of ‘San Giorgio dei Genovesi’, settled in the Historic Centre of Palermo (Italy). -

Hippocampus – Why Is It Studied So Frequently? Hipokampus – Zašto Se Toliko Prouþava?
Vojnosanit Pregl 2014; 71(2): 195–201. VOJNOSANITETSKI PREGLED Strana 195 UDC: 611.813:612.825 GENERAL REVIEW DOI: 10.2298/VSP130222043R Hippocampus – Why is it studied so frequently? Hipokampus – zašto se toliko prouþava? Veselin Radonjiü*, Slobodan Malobabiü†, Vidosava Radonjiü†, Laslo Puškaš†, Lazar Stijak†, Milan Aksiü†, Branislav Filipoviü† *Institute for Biocides and Medical Ecology, Belgrade, Serbia; †Institute of Anatomy, Faculty of Medicine, University of Belgrade, Belgrade, Serbia Key words: Kljuÿne reÿi: hippocampus; anatomy; neurophysiology; memory. hipokampus; anatomija; neurofiziologija; pamýenje. Introduction the medial wall of the temporal horn of the lateral ventricle. The name hippocampus to this structure was given by the From the very beginnings of brain research, the hippo- Bolognese anatomist Giulio Cesare Aranzio-Arantius in campus has been the focus of attention for anatomists. Nowa- 1564 which crossectional appearance resembled a seahorse 1 days, its complexity and clinical importance have attracted the to him. Winslow in 1732, used the term cornu arietis (ram`s interest of a vast number of researchers of different profiles. horn) for the appearance of the hippocampal section, which Work on hippocampal tissue facilitated some important De Garengeot in 1742 turned to cornu Ammonis after the neurophysiological discoveries: identification of excitatory Egyptian god Ammon who was depicted with a human body and inhibitory synapses, transmitters and receptors, discov- and the head of a ram with the horn7. The hippocampus ery of long-term potentiation and long-term depression, role proper is the more commonly used name for Ammon’s horn. of oscillations in neuronal networks, underlying mechanisms The name cornu Ammonis survived as the acronim CA for of epileptogenesis and of memory disorders 1.