Transcriptomic Profiling of Tumor-Infiltrating CD4+TIM-3+ T Cells Reveals Their Suppressive, Exhausted and Metastatic Characteristics In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Materials: Evaluation of Cytotoxicity and Α-Glucosidase Inhibitory Activity of Amide and Polyamino-Derivatives of Lupane Triterpenoids
Supplementary Materials: Evaluation of cytotoxicity and α-glucosidase inhibitory activity of amide and polyamino-derivatives of lupane triterpenoids Oxana B. Kazakova1*, Gul'nara V. Giniyatullina1, Akhat G. Mustafin1, Denis A. Babkov2, Elena V. Sokolova2, Alexander A. Spasov2* 1Ufa Institute of Chemistry of the Ufa Federal Research Centre of the Russian Academy of Sciences, 71, pr. Oktyabrya, 450054 Ufa, Russian Federation 2Scientific Center for Innovative Drugs, Volgograd State Medical University, Novorossiyskaya st. 39, Volgograd 400087, Russian Federation Correspondence Prof. Dr. Oxana B. Kazakova Ufa Institute of Chemistry of the Ufa Federal Research Centre of the Russian Academy of Sciences 71 Prospeсt Oktyabrya Ufa, 450054 Russian Federation E-mail: [email protected] Prof. Dr. Alexander A. Spasov Scientific Center for Innovative Drugs of the Volgograd State Medical University 39 Novorossiyskaya st. Volgograd, 400087 Russian Federation E-mail: [email protected] Figure S1. 1H and 13C of compound 2. H NH N H O H O H 2 2 Figure S2. 1H and 13C of compound 4. NH2 O H O H CH3 O O H H3C O H 4 3 Figure S3. Anticancer screening data of compound 2 at single dose assay 4 Figure S4. Anticancer screening data of compound 7 at single dose assay 5 Figure S5. Anticancer screening data of compound 8 at single dose assay 6 Figure S6. Anticancer screening data of compound 9 at single dose assay 7 Figure S7. Anticancer screening data of compound 12 at single dose assay 8 Figure S8. Anticancer screening data of compound 13 at single dose assay 9 Figure S9. Anticancer screening data of compound 14 at single dose assay 10 Figure S10. -

Analysis of Gene Expression Data for Gene Ontology
ANALYSIS OF GENE EXPRESSION DATA FOR GENE ONTOLOGY BASED PROTEIN FUNCTION PREDICTION A Thesis Presented to The Graduate Faculty of The University of Akron In Partial Fulfillment of the Requirements for the Degree Master of Science Robert Daniel Macholan May 2011 ANALYSIS OF GENE EXPRESSION DATA FOR GENE ONTOLOGY BASED PROTEIN FUNCTION PREDICTION Robert Daniel Macholan Thesis Approved: Accepted: _______________________________ _______________________________ Advisor Department Chair Dr. Zhong-Hui Duan Dr. Chien-Chung Chan _______________________________ _______________________________ Committee Member Dean of the College Dr. Chien-Chung Chan Dr. Chand K. Midha _______________________________ _______________________________ Committee Member Dean of the Graduate School Dr. Yingcai Xiao Dr. George R. Newkome _______________________________ Date ii ABSTRACT A tremendous increase in genomic data has encouraged biologists to turn to bioinformatics in order to assist in its interpretation and processing. One of the present challenges that need to be overcome in order to understand this data more completely is the development of a reliable method to accurately predict the function of a protein from its genomic information. This study focuses on developing an effective algorithm for protein function prediction. The algorithm is based on proteins that have similar expression patterns. The similarity of the expression data is determined using a novel measure, the slope matrix. The slope matrix introduces a normalized method for the comparison of expression levels throughout a proteome. The algorithm is tested using real microarray gene expression data. Their functions are characterized using gene ontology annotations. The results of the case study indicate the protein function prediction algorithm developed is comparable to the prediction algorithms that are based on the annotations of homologous proteins. -

Organ Level Protein Networks As a Reference for the Host Effects of the Microbiome
Downloaded from genome.cshlp.org on October 6, 2021 - Published by Cold Spring Harbor Laboratory Press 1 Organ level protein networks as a reference for the host effects of the microbiome 2 3 Robert H. Millsa,b,c,d, Jacob M. Wozniaka,b, Alison Vrbanacc, Anaamika Campeaua,b, Benoit 4 Chassainge,f,g,h, Andrew Gewirtze, Rob Knightc,d, and David J. Gonzaleza,b,d,# 5 6 a Department of Pharmacology, University of California, San Diego, California, USA 7 b Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California, San Diego, 8 California, USA 9 c Department of Pediatrics, and Department of Computer Science and Engineering, University of 10 California, San Diego California, USA 11 d Center for Microbiome Innovation, University of California, San Diego, California, USA 12 e Center for Inflammation, Immunity and Infection, Institute for Biomedical Sciences, Georgia State 13 University, Atlanta, GA, USA 14 f Neuroscience Institute, Georgia State University, Atlanta, GA, USA 15 g INSERM, U1016, Paris, France. 16 h Université de Paris, Paris, France. 17 18 Key words: Microbiota, Tandem Mass Tags, Organ Proteomics, Gnotobiotic Mice, Germ-free Mice, 19 Protein Networks, Proteomics 20 21 # Address Correspondence to: 22 David J. Gonzalez, PhD 23 Department of Pharmacology and Pharmacy 24 University of California, San Diego 25 La Jolla, CA 92093 26 E-mail: [email protected] 27 Phone: 858-822-1218 28 1 Downloaded from genome.cshlp.org on October 6, 2021 - Published by Cold Spring Harbor Laboratory Press 29 Abstract 30 Connections between the microbiome and health are rapidly emerging in a wide range of 31 diseases. -

Structural Characterization of the Human Eukaryotic Initiation Factor 3 Protein Complex by Mass Spectrometry*□S
Supplemental Material can be found at: http://www.mcponline.org/cgi/content/full/M600399-MCP200 /DC1 Research Structural Characterization of the Human Eukaryotic Initiation Factor 3 Protein Complex by Mass Spectrometry*□S Eugen Damoc‡, Christopher S. Fraser§, Min Zhou¶, Hortense Videler¶, Greg L. Mayeurʈ, John W. B. Hersheyʈ, Jennifer A. Doudna§, Carol V. Robinson¶**, and Julie A. Leary‡ ‡‡ Protein synthesis in mammalian cells requires initiation The initiation phase of eukaryotic protein synthesis involves factor eIF3, an ϳ800-kDa protein complex that plays a formation of an 80 S ribosomal complex containing the initi- Downloaded from central role in binding of initiator methionyl-tRNA and ator methionyl-tRNAi bound to the initiation codon in the mRNA to the 40 S ribosomal subunit to form the 48 S mRNA. This is a multistep process promoted by proteins initiation complex. The eIF3 complex also prevents pre- called eukaryotic initiation factors (eIFs).1 Currently at least 12 mature association of the 40 and 60 S ribosomal subunits eIFs, composed of at least 29 distinct subunits, have been and interacts with other initiation factors involved in start identified (1). Mammalian eIF3, the largest initiation factor, is a codon selection. The molecular mechanisms by which multisubunit complex with an apparent molecular mass of www.mcponline.org eIF3 exerts these functions are poorly understood. Since ϳ800 kDa. This protein complex plays an essential role in its initial characterization in the 1970s, the exact size, translation by binding directly to the 40 S ribosomal subunit composition, and post-translational modifications of and promoting formation of the 43 S preinitiation complex ⅐ ⅐ mammalian eIF3 have not been rigorously determined. -

Role of Mitochondrial Ribosomal Protein S18-2 in Cancerogenesis and in Regulation of Stemness and Differentiation
From THE DEPARTMENT OF MICROBIOLOGY TUMOR AND CELL BIOLOGY (MTC) Karolinska Institutet, Stockholm, Sweden ROLE OF MITOCHONDRIAL RIBOSOMAL PROTEIN S18-2 IN CANCEROGENESIS AND IN REGULATION OF STEMNESS AND DIFFERENTIATION Muhammad Mushtaq Stockholm 2017 All previously published papers were reproduced with permission from the publisher. Published by Karolinska Institutet. Printed by E-Print AB 2017 © Muhammad Mushtaq, 2017 ISBN 978-91-7676-697-2 Role of Mitochondrial Ribosomal Protein S18-2 in Cancerogenesis and in Regulation of Stemness and Differentiation THESIS FOR DOCTORAL DEGREE (Ph.D.) By Muhammad Mushtaq Principal Supervisor: Faculty Opponent: Associate Professor Elena Kashuba Professor Pramod Kumar Srivastava Karolinska Institutet University of Connecticut Department of Microbiology Tumor and Cell Center for Immunotherapy of Cancer and Biology (MTC) Infectious Diseases Co-supervisor(s): Examination Board: Professor Sonia Lain Professor Ola Söderberg Karolinska Institutet Uppsala University Department of Microbiology Tumor and Cell Department of Immunology, Genetics and Biology (MTC) Pathology (IGP) Professor George Klein Professor Boris Zhivotovsky Karolinska Institutet Karolinska Institutet Department of Microbiology Tumor and Cell Institute of Environmental Medicine (IMM) Biology (MTC) Professor Lars-Gunnar Larsson Karolinska Institutet Department of Microbiology Tumor and Cell Biology (MTC) Dedicated to my parents ABSTRACT Mitochondria carry their own ribosomes (mitoribosomes) for the translation of mRNA encoded by mitochondrial DNA. The architecture of mitoribosomes is mainly composed of mitochondrial ribosomal proteins (MRPs), which are encoded by nuclear genomic DNA. Emerging experimental evidences reveal that several MRPs are multifunctional and they exhibit important extra-mitochondrial functions, such as involvement in apoptosis, protein biosynthesis and signal transduction. Dysregulations of the MRPs are associated with severe pathological conditions, including cancer. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supplementary Figures 1-14 and Supplementary References
SUPPORTING INFORMATION Spatial Cross-Talk Between Oxidative Stress and DNA Replication in Human Fibroblasts Marko Radulovic,1,2 Noor O Baqader,1 Kai Stoeber,3† and Jasminka Godovac-Zimmermann1* 1Division of Medicine, University College London, Center for Nephrology, Royal Free Campus, Rowland Hill Street, London, NW3 2PF, UK. 2Insitute of Oncology and Radiology, Pasterova 14, 11000 Belgrade, Serbia 3Research Department of Pathology and UCL Cancer Institute, Rockefeller Building, University College London, University Street, London WC1E 6JJ, UK †Present Address: Shionogi Europe, 33 Kingsway, Holborn, London WC2B 6UF, UK TABLE OF CONTENTS 1. Supplementary Figures 1-14 and Supplementary References. Figure S-1. Network and joint spatial razor plot for 18 enzymes of glycolysis and the pentose phosphate shunt. Figure S-2. Correlation of SILAC ratios between OXS and OAC for proteins assigned to the SAME class. Figure S-3. Overlap matrix (r = 1) for groups of CORUM complexes containing 19 proteins of the 49-set. Figure S-4. Joint spatial razor plots for the Nop56p complex and FIB-associated complex involved in ribosome biogenesis. Figure S-5. Analysis of the response of emerin nuclear envelope complexes to OXS and OAC. Figure S-6. Joint spatial razor plots for the CCT protein folding complex, ATP synthase and V-Type ATPase. Figure S-7. Joint spatial razor plots showing changes in subcellular abundance and compartmental distribution for proteins annotated by GO to nucleocytoplasmic transport (GO:0006913). Figure S-8. Joint spatial razor plots showing changes in subcellular abundance and compartmental distribution for proteins annotated to endocytosis (GO:0006897). Figure S-9. Joint spatial razor plots for 401-set proteins annotated by GO to small GTPase mediated signal transduction (GO:0007264) and/or GTPase activity (GO:0003924). -

1 AGING Supplementary Table 2
SUPPLEMENTARY TABLES Supplementary Table 1. Details of the eight domain chains of KIAA0101. Serial IDENTITY MAX IN COMP- INTERFACE ID POSITION RESOLUTION EXPERIMENT TYPE number START STOP SCORE IDENTITY LEX WITH CAVITY A 4D2G_D 52 - 69 52 69 100 100 2.65 Å PCNA X-RAY DIFFRACTION √ B 4D2G_E 52 - 69 52 69 100 100 2.65 Å PCNA X-RAY DIFFRACTION √ C 6EHT_D 52 - 71 52 71 100 100 3.2Å PCNA X-RAY DIFFRACTION √ D 6EHT_E 52 - 71 52 71 100 100 3.2Å PCNA X-RAY DIFFRACTION √ E 6GWS_D 41-72 41 72 100 100 3.2Å PCNA X-RAY DIFFRACTION √ F 6GWS_E 41-72 41 72 100 100 2.9Å PCNA X-RAY DIFFRACTION √ G 6GWS_F 41-72 41 72 100 100 2.9Å PCNA X-RAY DIFFRACTION √ H 6IIW_B 2-11 2 11 100 100 1.699Å UHRF1 X-RAY DIFFRACTION √ www.aging-us.com 1 AGING Supplementary Table 2. Significantly enriched gene ontology (GO) annotations (cellular components) of KIAA0101 in lung adenocarcinoma (LinkedOmics). Leading Description FDR Leading Edge Gene EdgeNum RAD51, SPC25, CCNB1, BIRC5, NCAPG, ZWINT, MAD2L1, SKA3, NUF2, BUB1B, CENPA, SKA1, AURKB, NEK2, CENPW, HJURP, NDC80, CDCA5, NCAPH, BUB1, ZWILCH, CENPK, KIF2C, AURKA, CENPN, TOP2A, CENPM, PLK1, ERCC6L, CDT1, CHEK1, SPAG5, CENPH, condensed 66 0 SPC24, NUP37, BLM, CENPE, BUB3, CDK2, FANCD2, CENPO, CENPF, BRCA1, DSN1, chromosome MKI67, NCAPG2, H2AFX, HMGB2, SUV39H1, CBX3, TUBG1, KNTC1, PPP1CC, SMC2, BANF1, NCAPD2, SKA2, NUP107, BRCA2, NUP85, ITGB3BP, SYCE2, TOPBP1, DMC1, SMC4, INCENP. RAD51, OIP5, CDK1, SPC25, CCNB1, BIRC5, NCAPG, ZWINT, MAD2L1, SKA3, NUF2, BUB1B, CENPA, SKA1, AURKB, NEK2, ESCO2, CENPW, HJURP, TTK, NDC80, CDCA5, BUB1, ZWILCH, CENPK, KIF2C, AURKA, DSCC1, CENPN, CDCA8, CENPM, PLK1, MCM6, ERCC6L, CDT1, HELLS, CHEK1, SPAG5, CENPH, PCNA, SPC24, CENPI, NUP37, FEN1, chromosomal 94 0 CENPL, BLM, KIF18A, CENPE, MCM4, BUB3, SUV39H2, MCM2, CDK2, PIF1, DNA2, region CENPO, CENPF, CHEK2, DSN1, H2AFX, MCM7, SUV39H1, MTBP, CBX3, RECQL4, KNTC1, PPP1CC, CENPP, CENPQ, PTGES3, NCAPD2, DYNLL1, SKA2, HAT1, NUP107, MCM5, MCM3, MSH2, BRCA2, NUP85, SSB, ITGB3BP, DMC1, INCENP, THOC3, XPO1, APEX1, XRCC5, KIF22, DCLRE1A, SEH1L, XRCC3, NSMCE2, RAD21. -

Mouse MRPL17 ORF Mammalian Expression Plasmid, N-Gfpspark Tag
Mouse MRPL17 ORF mammalian expression plasmid, N-GFPSpark tag Catalog Number: MG53556-ANG General Information Plasmid Resuspension protocol Gene : mitochondrial ribosomal protein L17 1. Centrifuge at 5,000×g for 5 min. Official Symbol : MRPL17 2. Carefully open the tube and add 100 l of sterile water to Synonym : Rpml26; MRP-L26 dissolve the DNA. Source : Mouse 3. Close the tube and incubate for 10 minutes at room cDNA Size: 531bp temperature. RefSeq : NM_025301.2 4. Briefly vortex the tube and then do a quick spin to Description concentrate the liquid at the bottom. Speed is less than Lot : Please refer to the label on the tube 5000×g. Vector : pCMV3-N-GFPSpark 5. Store the plasmid at -20 ℃. Shipping carrier : Each tube contains approximately 10 μg of lyophilized plasmid. The plasmid is ready for: Storage : • Restriction enzyme digestion The lyophilized plasmid can be stored at ambient temperature for three months. • PCR amplification Quality control : • E. coli transformation The plasmid is confirmed by full-length sequencing with primers • DNA sequencing in the sequencing primer list. Sequencing primer list : E.coli strains for transformation (recommended but not limited) pCMV3-F: 5’ CAGGTGTCCACTCCCAGGTCCAAG 3’ Most commercially available competent cells are appropriate for pcDNA3-R : 5’ GGCAACTAGAAGGCACAGTCGAGG 3’ the plasmid, e.g. TOP10, DH5α and TOP10F´. Or Forward T7 : 5’ TAATACGACTCACTATAGGG 3’ ReverseBGH : 5’ TAGAAGGCACAGTCGAGG 3’ pCMV3-F and pcDNA3-R are designed by Sino Biological Inc. Customers can order the primer pair from any oligonucleotide supplier. Manufactured By Sino Biological Inc., FOR RESEARCH USE ONLY. NOT FOR USE IN HUMANS. Fax :+86-10-51029969 Tel:+86- 400-890-9989 http://www.sinobiological.com Mouse MRPL17 ORF mammalian expression plasmid, N-GFPSpark tag Catalog Number: MG53556-ANG Vector Information All of the pCMV vectors are designed for high-level stable and transient expression in mammalian hosts. -
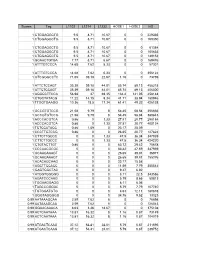
Scores Tag L1102 L1214 L1232 HOSE1 HOSE2 HS 1
Scores Tag L1102 L1214 L1232 HOSE1 HOSE2 HS 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 229335 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 169350 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 61384 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 105633 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 149152 1 GCAACTGTGA 7.77 8.71 6.67 0 0 169476 1 ATTTGTCCCA 14.68 7.62 5.33 0 0 57301 1 ATTTGTCCCA 14.68 7.62 5.33 0 0 356122 1 GTCGGGCCTC 71.65 39.18 22.67 1.16 0 73769 1 ATTCTCCAGT 35.39 39.18 44.01 85.74 89.13 458218 1 ATTCTCCAGT 35.39 39.18 44.01 85.74 89.13 406300 1 AGGGCTTCCA 56.98 37 69.35 134.4 141.05 458148 1 CTGCTATACG 11.22 14.15 9.34 41.71 38.94 180946 1 TTGGTGAAGG 10.36 18.5 17.34 61.41 49.32 426138 1 GCCGTGTCCG 21.58 9.79 8 54.45 58.84 356666 1 GCCGTGTCCG 21.58 9.79 8 54.45 58.84 380843 1 ACCCACGTCA 0.86 0 1.33 27.81 20.77 298184 1 ACCCACGTCA 0.86 0 1.33 27.81 20.77 400124 1 TCTCCATACC 0.86 1.09 0 23.17 25.09 1 CCCTTGTCCG 0.86 0 0 26.65 20.77 127824 1 CTTCTTGCCC 0 0 1.33 47.5 36.34 347939 1 CTTCTTGCCC 0 0 1.33 47.5 36.34 424220 1 CTGTACTTGT 0.86 0 0 63.72 29.42 75678 1 CCCAACGCGC 0 0 0 83.42 47.59 347939 1 GCAAGAAAGT 0 0 0 26.65 39.81 36977 1 GCAAGAAAGT 0 0 0 26.65 39.81 155376 1 ACACAGCAAG 0 0 0 23.17 15.58 1 AGCTTCCACC 0 0 0 11.59 7.79 355542 1 GAGTGGCTAC 0 0 0 9.27 6.92 1 ATGGTGGGGG 0 0 0 8.11 22.5 343586 1 AGATCCCAAG 0 0 0 5.79 8.65 50813 1 TGGAAGGAGG 0 0 0 8.11 6.06 1 TAGCCGGGAC 0 0 0 5.79 7.79 107740 1 TGTGGATGTG 0 0 0 4.63 12.11 180878 1 GGGTAGGGGG 0 0 0 34.76 9.52 13323 0.99 AATAAAGCAA 2.59 7.62 8 0 0 76698 0.99 AATAAAGCAA 2.59 7.62 8 0 0 126043 0.99 GGAACAAACA 8.63 3.26 18.67 0 0 375108 -

Systematically Profiling the Expression of Eif3 Subunits in Glioma Reveals
Chai et al. Cancer Cell Int (2019) 19:155 https://doi.org/10.1186/s12935-019-0867-1 Cancer Cell International PRIMARY RESEARCH Open Access Systematically profling the expression of eIF3 subunits in glioma reveals the expression of eIF3i has prognostic value in IDH-mutant lower grade glioma Rui‑Chao Chai1,4,6†, Ning Wang2†, Yu‑Zhou Chang3, Ke‑Nan Zhang1,6, Jing‑Jun Li1,6, Jun‑Jie Niu5, Fan Wu1,6*, Yu‑Qing Liu1,6* and Yong‑Zhi Wang1,3,4,6* Abstract Background: Abnormal expression of the eukaryotic initiation factor 3 (eIF3) subunits plays critical roles in tumo‑ rigenesis and progression, and also has potential prognostic value in cancers. However, the expression and clinical implications of eIF3 subunits in glioma remain unknown. Methods: Expression data of eIF3 for patients with gliomas were obtained from the Chinese Glioma Genome Atlas (CGGA) (n 272) and The Cancer Genome Atlas (TCGA) (n 595). Cox regression, the receiver operating characteristic (ROC) curves= and Kaplan–Meier analysis were used to study= the prognostic value. Gene oncology (GO) and gene set enrichment analysis (GSEA) were utilized for functional prediction. Results: In both the CGGA and TCGA datasets, the expression levels of eIF3d, eIF3e, eIF3f, eIF3h and eIF3l highly were associated with the IDH mutant status of gliomas. The expression of eIF3b, eIF3i, eIF3k and eIF3m was increased with the tumor grade, and was associated with poorer overall survival [All Hazard ratio (HR) > 1 and P < 0.05]. By contrast, the expression of eIF3a and eIF3l was decreased in higher grade gliomas and was associated with better overall sur‑ vival (Both HR < 1 and P < 0.05). -

Using Edsurvey to Analyze NCES Data: an Illustration of Analyzing NAEP Primer
Using EdSurvey to Analyze NCES Data: An Illustration of Analyzing NAEP Primer Developed by Michael Lee, Paul Bailey, Ahmad Emad, Ting Zhang, Trang Nguyen, and Jiao Yu*† February 21, 2020 Overview of the EdSurvey Package National Assessment of Educational Progress (NAEP) datasets from the National Center for Education Statistics (NCES) require special statistical methods to analyze. Because of their scope and complexity, the EdSurvey package gives users functions to perform analyses that account for both complex sample survey designs and the use of plausible values. The EdSurvey package also seamlessly takes advantage of the LaF package to read in data only when required for an analysis. Users with computers that have insuÿcient memory to read in entire NAEP datasets can still do analyses without having to write special code to read in just the appropriate variables. This situation is addressed directly in the EdSurvey package—behind the scenes and without any special tuning by the user. Vignette Outline This vignette will describe the basics of using the EdSurvey package for analyzing NAEP data as follows. • Notes – Additional resources – Vignette notation – Software requirements • Setting up the environment for analyzing NCES data – Installing and loading EdSurvey – Philosophy of Conducting Analyses Using the EdSurvey Package – Downloading data – Reading in data – Getting to know the data format – Removing special values • Explore Variable Distributions with summary2 • Subsetting the data • Retrieving data for further manipulation with getData *This publication was prepared for NCES under Contract No. ED-IES-12-D-0002 with the American Institutes for Research. Mention of trade names, commercial products, or organizations does not imply endorsement by the U.S.