Symbiosome Development Have Been Indicated
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Transport in Plants and in Root Nodules
DEVELOPMENT OF SPECIALIZED TRANSPORT PATHWAYS IN PISUM SATIVUM ROOT NODULES by Cherish A. Warner A dissertation submitted to the Faculty of the University of Delaware in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biological Sciences Fall 2018 © 2018 Cherish A. Warner All Rights Reserved DEVELOPMENT OF SPECIALIZED TRANSPORT PATHWAYS IN PISUM SATIVUM ROOT NODULES by Cherish A. Warner Approved: __________________________________________________________ E. Fidelma Boyd, Ph.D. Chair of the Department of Biological Sciences Approved: __________________________________________________________ John Pelesko, Ph.D. Interim Dean of the College of Arts and Sciences Approved: __________________________________________________________ Douglas J. Doren, Ph.D. Interim Vice Provost for Graduate and Professional Education I certify that I have read this dissertation and that in my opinion it meets the academic and professional standard required by the University as a dissertation for the degree of Doctor of Philosophy. Signed: __________________________________________________________ D. Janine Sherrier, Ph.D. Professor in charge of dissertation I certify that I have read this dissertation and that in my opinion it meets the academic and professional standard required by the University as a dissertation for the degree of Doctor of Philosophy. Signed: __________________________________________________________ Jeffrey L. Caplan, Ph.D. Member of dissertation committee I certify that I have read this dissertation and that in my opinion it meets the academic and professional standard required by the University as a dissertation for the degree of Doctor of Philosophy. Signed: __________________________________________________________ Diane Herson, Ph.D. Member of dissertation committee I certify that I have read this dissertation and that in my opinion it meets the academic and professional standard required by the University as a dissertation for the degree of Doctor of Philosophy. -

Types of Alcoholic Beverages and Blood Lipids in a French Population
24 J Epidemiol Community Health: first published as 10.1136/jech.56.1.24 on 1 January 2002. Downloaded from RESEARCH REPORT Types of alcoholic beverages and blood lipids in a French population J-B Ruidavets, P Ducimetière, D Arveiler, P Amouyel, A Bingham, A Wagner, D Cottel, B Perret, J Ferrières ............................................................................................................................. J Epidemiol Community Health 2002;56:24–28 Study objective: Prospective studies have shown a consistent relation between alcohol consumption and decreasing incidence of coronary artery disease. The protective effect of alcohol could be medi- ated through increased levels of HDL cholesterol (HDL-c). The aim of this study was to examine the rela- tion between blood lipid levels and the consumption of different types of alcoholic beverages among 1581 men and 1535 women. See end of article for Design: Data from representative cross sectional surveys (1994–1997) in three different regions of authors’ affiliations France were used. The consumption of the different types of alcohol was quantified using a recall ....................... method according to a typical weekly consumption. Correspondence: Dr J-B Main results: The median daily alcohol intake was 24 g for men and 4 g for women. After adjustment Ruidavets, INSERM U558, for confounders, total alcohol showed a positive and significant association with HDL-c and triglycerides Département (TG) in both sexes. In multivariate analysis, wine was positively associated with HDL-c. Beer was posi- d’épidémiologie, Faculté tively associated with HDL-c in men and with triglycerides in men and women. When taking drinking de médecine, 37, allées Jules Guesde, 31073 patterns into account, wine drinkers had higher HDL-c levels than non-wine drinkers. -

Updated Checklist of Marine Fishes (Chordata: Craniata) from Portugal and the Proposed Extension of the Portuguese Continental Shelf
European Journal of Taxonomy 73: 1-73 ISSN 2118-9773 http://dx.doi.org/10.5852/ejt.2014.73 www.europeanjournaloftaxonomy.eu 2014 · Carneiro M. et al. This work is licensed under a Creative Commons Attribution 3.0 License. Monograph urn:lsid:zoobank.org:pub:9A5F217D-8E7B-448A-9CAB-2CCC9CC6F857 Updated checklist of marine fishes (Chordata: Craniata) from Portugal and the proposed extension of the Portuguese continental shelf Miguel CARNEIRO1,5, Rogélia MARTINS2,6, Monica LANDI*,3,7 & Filipe O. COSTA4,8 1,2 DIV-RP (Modelling and Management Fishery Resources Division), Instituto Português do Mar e da Atmosfera, Av. Brasilia 1449-006 Lisboa, Portugal. E-mail: [email protected], [email protected] 3,4 CBMA (Centre of Molecular and Environmental Biology), Department of Biology, University of Minho, Campus de Gualtar, 4710-057 Braga, Portugal. E-mail: [email protected], [email protected] * corresponding author: [email protected] 5 urn:lsid:zoobank.org:author:90A98A50-327E-4648-9DCE-75709C7A2472 6 urn:lsid:zoobank.org:author:1EB6DE00-9E91-407C-B7C4-34F31F29FD88 7 urn:lsid:zoobank.org:author:6D3AC760-77F2-4CFA-B5C7-665CB07F4CEB 8 urn:lsid:zoobank.org:author:48E53CF3-71C8-403C-BECD-10B20B3C15B4 Abstract. The study of the Portuguese marine ichthyofauna has a long historical tradition, rooted back in the 18th Century. Here we present an annotated checklist of the marine fishes from Portuguese waters, including the area encompassed by the proposed extension of the Portuguese continental shelf and the Economic Exclusive Zone (EEZ). The list is based on historical literature records and taxon occurrence data obtained from natural history collections, together with new revisions and occurrences. -

Iron Transport Across Symbiotic Membranes of Nitrogen-Fixing Legumes
International Journal of Molecular Sciences Review Iron Transport across Symbiotic Membranes of Nitrogen-Fixing Legumes David A. Day 1,* and Penelope M. C. Smith 2 1 College of Science and Engineering, Flinders University, GPO Box 2100, Adelaide, SA 5001, Australia 2 School of Life Sciences, La Trobe University, Melbourne, VIC 3083, Australia; [email protected] * Correspondence: David.Day@flinders.edu.au Abstract: Iron is an essential nutrient for the legume-rhizobia symbiosis and nitrogen-fixing bac- teroids within root nodules of legumes have a very high demand for the metal. Within the infected cells of nodules, the bacteroids are surrounded by a plant membrane to form an organelle-like structure called the symbiosome. In this review, we focus on how iron is transported across the symbiosome membrane and accessed by the bacteroids. Keywords: legumes; nitrogen fixation; symbiosomes; iron 1. Introduction Iron is an essential nutrient for cell function and plants have developed specialised mechanisms for mobilising, absorbing and storing the metal. Cellular iron homeostasis is important because iron is toxic when present in excess. Symbiotically grown legumes have a particularly high requirement for iron and low iron severely retards their growth and their ability to fix atmospheric nitrogen [1,2]. Legumes form a symbiosis with nitrogen-fixing soil bacteria (rhizobia) that enables the plants to utilize atmospheric nitrogen for growth. Infection of the legume root by rhizobia Citation: Day, D.A.; Smith, P.M.C. results in the formation of specialized organs called nodules that provide the microaerobic Iron Transport across Symbiotic conditions required for operation of the bacterium’s nitrogenase enzyme. -

Special Feature
Ecology, 84(4), 2003, pp. 858±868 q 2003 by the Ecological Society of America MOLECULAR SIGNALS AND RECEPTORS: CONTROLLING RHIZOSPHERE INTERACTIONS BETWEEN PLANTS AND OTHER ORGANISMS ANN M. HIRSCH,1,7 W. D IETZ BAUER,2 DAVID M. BIRD,3 JULIE CULLIMORE,4 BRETT TYLER,5 AND JOHN I. YODER6 1Department of Molecular, Cell and Developmental Biology, University of California, Los Angeles, California 90095 USA 2Department of Horticulture and Crop Science, Ohio State University, Columbus, Ohio 43210 USA 3Department of Plant Pathology, North Carolina State University, Raleigh, North Carolina 27695 USA 4Laboratoire de Biologie MoleÂculaire des Relations Plantes-Microorganismes, CNRS-INRA BP27, 31326 Castanet-Tolosan Cedex, France 5Virginia Bioinformatics Institute, 1880 Pratt Drive, Blacksburg, Virginia 24061 USA 6Department of Vegetable Crops, University of California, Davis, California 95616 USA Abstract. Rhizosphere interactions are affected by many different regulatory signals. As yet, however, only a few have been identi®ed. Signals, by de®nition, contain information, react with a receptor, and elicit a response. Signals may thus represent the highest level of evolved response in rhizosphere communities and, in that sense, occupy a supreme control point. At the same time, some signals may function as modulators of downstream responses, rather than on/off switches. To assess these possibilities, several interactions between plants and soil organisms are described, starting with the molecular interactions between legu- minous plants and symbiotic bacteria of the family Rhizobiaceae, one of the best-charac- terized plant±microbe associations in the rhizosphere. We then examine other interactions between plants and soil organisms for overlap and/or connections with the rhizosphere signals utilized in the legume±Rhizobium symbiosis. -

Beverage Formula Seminar
BEVERAGE FORMULA SEMINAR Formulation Team Advertising, Labeling and Formulation Division TTB May 3, 2006 AGENDA • Advertising, Labeling & Formulation Division (ALFD) • Basics of TTB Formulation • Wine • Distilled Spirits • Malt Beverage WHERE DOES ALFD FIT IN TTB? John Manfreda Administrator Vicky I. McDowell Deputy Administrator Cheri Mitchell Bill Foster Mary Ryan Assistant Administrator Assistant Administrator Assistant Administrator (Management) (Headquarters Operations) (Field Operations) Advertising Labeling and National Revenue Formulation Division Center Regulations and Rulings Tax Audit Division Division International Trade Trade Investigations Division Division Scientific Services Division Advertising, Labeling and Formulation Division Division Director ALFD Karen Freelove (202) 927-8087 Technical Advisor Division Admin. Asst. Ed Reisman Joyce Rose (202) 927-8485 Assistant Director Assistant Director Supervisory Mgmt Assistant Director Teresa Knapp Vacant Analyst Susan Weil Wine Labeling Market Compliance Donna Smith Formulation/DS&MB Office Office Info. Tech Office Labeling Offices (202) 927-1975 (202) 927-8136 (202) 927-8107 (202) 927-8122 Customer Service Program Manager Program Analysts Program Manager Specialists 1 2 1 2 Customer Service Formula Specialists Market Compliance Specialist 3 QA Specialists Specialists 1 2 5 (one vacancy) QA Specialist ITT Specialist 1 Label Specialists 1 Customer Service 1 1 Clerks Specialist 3 (one vacancy) 1 Administrative Asst. 1 Label Specialists 3 ALFD Contact Information • Mailing Address -

Contents of Volume 42 (2012)
CONTENTS OF VOLUME 42 (2012) Issue 1 Tripp-Valdez A., Arreguín-Sánchez F., Zetina-Rejón M.J. The food of Selene peruviana (Actinopterygii: Perciformes: Carangidae) in the southern Gulf of California .................. 1 Zarrad R., Alemany F., Jarboui O., Garcia A., Akrout F. Comparative characterization of the spawning environments of European anchovy, Engraulis encrasicolus , and round sardinella, Sardinella aurita (Actinopterygii: Clupeiformes) in the eastern coast of Tunisia ............................... 9 Ordines F., Valls M., Gouraguine A. Biology, feeding, and habitat preferences of Cadenat’s rockfish, Scorpaena loppei (Actinopterygii: Scorpaeniformes: Scorpaenidae), in the Balearic Islands (western Mediterranean) ..................................... 21 Socha M., Sokołowska-Mikołajczyk M., Szczerbik P., Chyb J., Mikołajczyk T., Epler P. The effect of polychlorinated biphenyls mixture (Aroclor 1254) on the embryonic development and hatching of Prussian carp, Carassius gibelio , and common carp, Cyprinus carpio (Actinopterygii: Cypriniformes: Cyprinidae) ............................................................................................................................................................... 31 Khan A., Ghosh K. Characterization and identification of gut-associated phytase-producing bacteria in some fresh water fish cultured in ponds ...................................................................................................................................................................................... -

Cellular and Molecular Aspects of Cnidarian-Algal Associations
AN ABSTRACT OF THE THESIS OF Jodi A. Schwarz for the degree of Doctor of Philosophy in Zoology presented on October 18, 2002. Title: Cellular and Molecular Aspects of Cnidarian-Algal Associations Redacted for Privacy Abstract approved: Virginia M. Weis Intracellular symbioses between cnidarians and dinoflagellates from the genus Symbiodinium are widespread throughout the marine environment. These associations are ecologically significant, especially in tropical waters where symbiotic interactions between corals and Symbiodinium culminate in the formation of limestone reefs. This thesis focuses on cellular and molecular aspects of the symbiosis, specifically the initiation of the symbiosis and characterization of a host gene, sym32, that is believed to function in the symbiosis. Sym32 was originally identified as a differentially expressed protein in symbiotic vs. aposymbiotic individuals of the sea anemone, Anthopleura elegantissima. Based on its deduced amino acid sequence, sym32 belongs to a family of cell adhesion proteins that play roles in cell recognition in a diverse array of organisms. Chapter 2 examines the process by which a new cnidarian host acquires its first symbionts. Larvae of the scleractinian coral Fungia scutaria, which are initially aposymbiotic, acquired symbionts while feeding. Symbionts that entered the larval gastric cavity with food were subsequently taken into host gastrodermal cells by phagocytosis. Chapter 3 describes immunolocalization of sym32 in A. elegantissima tentacles. In aposymbiotic tentacles, sym32 was localized to vesicles within the host gastrodermal cells. Symbiotic tentacles lacked sym32-containing vesicles. Instead, sym32 was present among the membranes that enclose the symbionts within host cells. Western blots of proteins fromSymbiodiniumrevealed a 45/48kD doublet that cross-reacts with anti-sym32 antiserum. -
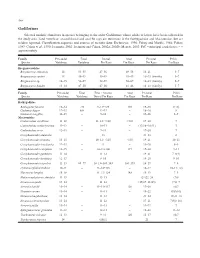
Gadiformes Selected Meristic Characters in Species Belonging to the Order Gadiformes Whose Adults Or Larvae Have Been Collected in the Study Area
548 Gadiformes Selected meristic characters in species belonging to the order Gadiformes whose adults or larvae have been collected in the study area. Total vertebrae, second dorsal and anal fin rays are numerous in the Bathygadidae and Macrouridae, but are seldom reported. Classification sequence and sources of meristic data: Eschmeyer, 1990; Fahay and Markle, 1984; Fahay, 1989; Cohen et al., 1990; Iwamoto, 2002; Iwamoto and Cohen, 2002a; 2002b; Merrett, 2003. PrC = principal caudal rays; ~ = approximately Family Precaudal Total Dorsal Anal Pectoral Pelvic Species Vertebrae Vertebrae Fin Rays Fin Rays Fin Rays Fin Rays Bregmacerotidae Bregmaceros atlanticus 14 53–55 47–56 49–58 16–21 5–7 Bregmaceros cantori 14 45–49 45–49 45–49 16–23 (family) 5–7 Bregmaceros sp. 14–15 52–59 52–59 58–69 16–23 (family) 5–7 Bregmaceros houdei 13–14 47–50 47–50 41–46 16–23 (family) 5–7 Family Precaudal Total First + Second Anal Pectoral Pelvic Species Vertebrae Vertebrae Dorsal Fin Rays Fin Rays Fin Rays Fin Rays Bathygadidae Bathygadus favosus 12–14 ~70 9–11+125 110 15–18 9(10) Gadomus dispar 12–13 80+ 12–13 – 18–20 8 Gadomus longifilis 11–13 – 9–11 – 14–16 8–9 Macrouridae Caelorinchus caribbeus 11–12 – 11–12+>110 >110 17–20 7 Caelorinchus coelorhynchus 11–12 – 10–11 – (17)18–20(21) 7 Caelorinchus occa 12–13 – 9–11 – 17–20 7 Coryphaenoides alateralis – 13 – 21–23 8 Coryphaenoides armatus 13–15 – 10–12+~125 ~135 19–21 10–11 Coryphaenoides brevibarbis 12–13 – 9 – 19–20 8–9 Coryphaenoides carapinus 12–15 – 10–11+100 117 17–20 9–11 Coryphaenoides guentheri -

Excise the World of Intoxication
REVENUE EARNING DEPARTMENTS - EXCISE THE WORLD OF INTOXICATION Alcoholic Drinks: Previous Era Alcoholic Drinks: History Alcoholic drinks made from fermented food stuffs have been in used from ancient times. Fermented drinks antedate distilled spirits, though the process of distillation was known to the ancient Assyrians, Chinese, Greeks and Hindus. The manufacture, sale and consumption of intoxicating liquor have been subject to state control from very early times in India. Alcoholic Drinks - in India Drinks were known in India in Vedik and Post Vedik times. The celestial drink of Vedik period is known as Soma. • Sura is fermented beverage during Athavana Veda period. Alcoholic Drinks – Making in different periods • Pulasty’s • Kautilya’s Alcohol making : Pulasty’s Period • Panasa( Liquor from Jack fruit) • Madhvika (Mohowa Liquor) • Draksha (Liquor from Grape) • Saira (Long pepper Liquor) • Madhuka (Honey Liquor) • Arishta (Soap Berry Liquor) • Khajura (Date Liquor) • Maireya (Rum) • Tala (Palm Liquor) • Narikelaja (Coconut Liquor) • Sikhshava (Cane Liquor) • Sura / Arrack. Alcohol making : Kautilya’s Period • Medaka • Prasanna • Asava • Arisha • Maireya • Madhu Indian Alcoholic Beverages Indian Alcoholic Beverages : Types • Traditional Alcoholic Beverages • Non- Traditional Alcoholic Beverages Traditional Alcoholic Beverages • Feni • Hudamaba • Palm Wine • Handia • Hariya • Kaidum • Desidaru • Sonti • Kodo Kojaanr • Apo / Apung • Sulai • Laopani • Arrack • Sundakanji • Luqdi • Bangla • Sura • Mahua • Bitchi • Tati Kallu • Mahuli • Chhaang • Tharra • Mandia Pej • Cholai • Zawlaidi • Manri • Chuak • Zutho • Pendha • Sekmai Non - Traditional Alcoholic Beverages • Indian Beer • Indian Brandy • Indian made Foreign Liquor • Indian Rum • Indian Vodka • Indian Wine Alcoholic Beverages Alcohol Beverages : as a source of Revenue Alcoholic beverages received to distinctions with the advent of the British Rule in India. -

Genetic Diversity of Antarctic Fish
GENETIC DIVERSITY OF ANTARCTIC FISH Elaine M. Fitzcharles A Thesis Submitted for the Degree of PhD at the University of St Andrews 2014 Full metadata for this item is available in Research@StAndrews:FullText at: http://research-repository.st-andrews.ac.uk/ Please use this identifier to cite or link to this item: http://hdl.handle.net/10023/6860 This item is protected by original copyright Genetic Diversity of Antarctic Fish Elaine M. Fitzcharles This thesis is submitted in partial fulfilment for the degree of PhD at the University of St Andrews May 2014 Supervisor of studies Prof. Alex Rogers (University of Oxford) Dr Melody Clark (British Antarctic Survey) Prof. Andrew Brierley (University of St Andrews) Sponsoring establishment British Antarctic Survey Natural Environment Research Council High Cross Madingley Road Cambridge CB3 0ET United Kingdom ii DECLARATIONS 1. Candidate’s declarations: I, Elaine Fitzcharles, hereby certify that this thesis, which is approximately 35000 words in length, has been written by me, and that it is the record of work carried out by me, or principally by myself in collaboration with others as acknowledged, and that it has not been submitted in any previous application for a higher degree. I was admitted as a research student in September 2005 and as a candidate for the degree of Doctor of Philosophy in April 2007; the higher study for which this is a record was carried out in the University of St Andrews between 2005 and 2014. Date 23/5/11 Signature of candidate …………………….............. 2. Supervisor’s declaration: I hereby certify that the candidate has fulfilled the conditions of the Resolution and Regulations appropriate for the degree of Doctor of Philosophy in the University of St Andrews and that the candidate is qualified to submit this thesis in application for that degree. -

Lifestyle Adaptations of Rhizobium from Rhizosphere to Symbiosis
Lifestyle adaptations of Rhizobium from rhizosphere to symbiosis Rachel M. Wheatleya,1, Brandon L. Forda,1,LiLib,1, Samuel T. N. Aroneya, Hayley E. Knightsa, Raphael Ledermanna, Alison K. Easta, Vinoy K. Ramachandrana,2, and Philip S. Poolea,2 aDepartment of Plant Sciences, University of Oxford, OX1 3RB Oxford, United Kingdom; and bChinese Academy of Sciences Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture, Wuhan Botanical Garden, Chinese Academy of Sciences, 430074 Wuhan, People’s Republic of China Edited by Éva Kondorosi, Hungarian Academy of Sciences, Biological Research Centre, Szeged, Hungary, and approved August 4, 2020 (received for review May 7, 2020) By analyzing successive lifestyle stages of a model Rhizobium– nodule cells and undergo terminal differentiation into N2-fixing legume symbiosis using mariner-based transposon insertion se- bacteroids (10). Nodules provide a protective microaerobic envi- quencing (INSeq), we have defined the genes required for rhizo- ronment to maintain oxygen-labile nitrogenase (6). In exchange + sphere growth, root colonization, bacterial infection, N2-fixing for NH4 and alanine, the legume host provides carbon sources to bacteroids, and release from legume (pea) nodules. While only 27 fuel this process, primarily as dicarboxylic acids (13, 14). genes are annotated as nif and fix in Rhizobium leguminosarum,we However, nodule infection is only one stage of the lifestyle of show 603 genetic regions (593 genes, 5 transfer RNAs, and 5 RNA rhizobia, and they spend much of their time surviving in the rhi- features) are required for the competitive ability to nodulate pea and zosphere, the zone of soil immediately surrounding roots (15).