Diptera: Chironomidae: Diamesinae) from the Mountains of Central Asia and the Middle East, with Description and DNA Barcoding of New Taxa
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

ARTHROPOD COMMUNITIES and PASSERINE DIET: EFFECTS of SHRUB EXPANSION in WESTERN ALASKA by Molly Tankersley Mcdermott, B.A./B.S
Arthropod communities and passerine diet: effects of shrub expansion in Western Alaska Item Type Thesis Authors McDermott, Molly Tankersley Download date 26/09/2021 06:13:39 Link to Item http://hdl.handle.net/11122/7893 ARTHROPOD COMMUNITIES AND PASSERINE DIET: EFFECTS OF SHRUB EXPANSION IN WESTERN ALASKA By Molly Tankersley McDermott, B.A./B.S. A Thesis Submitted in Partial Fulfillment of the Requirements for the Degree of Master of Science in Biological Sciences University of Alaska Fairbanks August 2017 APPROVED: Pat Doak, Committee Chair Greg Breed, Committee Member Colleen Handel, Committee Member Christa Mulder, Committee Member Kris Hundertmark, Chair Department o f Biology and Wildlife Paul Layer, Dean College o f Natural Science and Mathematics Michael Castellini, Dean of the Graduate School ABSTRACT Across the Arctic, taller woody shrubs, particularly willow (Salix spp.), birch (Betula spp.), and alder (Alnus spp.), have been expanding rapidly onto tundra. Changes in vegetation structure can alter the physical habitat structure, thermal environment, and food available to arthropods, which play an important role in the structure and functioning of Arctic ecosystems. Not only do they provide key ecosystem services such as pollination and nutrient cycling, they are an essential food source for migratory birds. In this study I examined the relationships between the abundance, diversity, and community composition of arthropods and the height and cover of several shrub species across a tundra-shrub gradient in northwestern Alaska. To characterize nestling diet of common passerines that occupy this gradient, I used next-generation sequencing of fecal matter. Willow cover was strongly and consistently associated with abundance and biomass of arthropods and significant shifts in arthropod community composition and diversity. -

Genomanalyse Von Prodiamesa Olivacea
Genomanalyse von Prodiamesa olivacea Dissertation zur Erlangung des Grades Doktor der Naturwissenschaften (Dr. rer. nat.) am Fachbereich Biologie der Johannes Gutenberg-Universität in Mainz Sarah Brunck geb. 08.08.1987 in Mainz Mainz, 2016 Dekan: 1. Berichterstatter: 2. Berichterstatter: Tag der mündlichen Prüfung: ii Inhaltsverzeichnis Inhaltsverzeichnis ................................................................................................................................ iii 1 Einleitung ........................................................................................................................................... 1 1.1 Die Familie der Chironomiden ................................................................................................. 1 1.1.1 Die Gattung Chironomus ..................................................................................................... 3 1.1.2 Die Gattung Prodiamesa ....................................................................................................... 6 1.2 Die Struktur von Insekten-Genomen am Beispiel der Chironomiden ............................... 9 1.2.1 Hochrepetitive DNA-Sequenzen ..................................................................................... 11 1.2.2 Mittelrepetitive DNA-Sequenzen bzw. Gen-Familien ................................................. 13 1.2.3 Gene und genregulatorische Sequenzen ........................................................................ 17 1.3 Zielsetzung ............................................................................................................................... -

Ohio EPA Macroinvertebrate Taxonomic Level December 2019 1 Table 1. Current Taxonomic Keys and the Level of Taxonomy Routinely U
Ohio EPA Macroinvertebrate Taxonomic Level December 2019 Table 1. Current taxonomic keys and the level of taxonomy routinely used by the Ohio EPA in streams and rivers for various macroinvertebrate taxonomic classifications. Genera that are reasonably considered to be monotypic in Ohio are also listed. Taxon Subtaxon Taxonomic Level Taxonomic Key(ies) Species Pennak 1989, Thorp & Rogers 2016 Porifera If no gemmules are present identify to family (Spongillidae). Genus Thorp & Rogers 2016 Cnidaria monotypic genera: Cordylophora caspia and Craspedacusta sowerbii Platyhelminthes Class (Turbellaria) Thorp & Rogers 2016 Nemertea Phylum (Nemertea) Thorp & Rogers 2016 Phylum (Nematomorpha) Thorp & Rogers 2016 Nematomorpha Paragordius varius monotypic genus Thorp & Rogers 2016 Genus Thorp & Rogers 2016 Ectoprocta monotypic genera: Cristatella mucedo, Hyalinella punctata, Lophopodella carteri, Paludicella articulata, Pectinatella magnifica, Pottsiella erecta Entoprocta Urnatella gracilis monotypic genus Thorp & Rogers 2016 Polychaeta Class (Polychaeta) Thorp & Rogers 2016 Annelida Oligochaeta Subclass (Oligochaeta) Thorp & Rogers 2016 Hirudinida Species Klemm 1982, Klemm et al. 2015 Anostraca Species Thorp & Rogers 2016 Species (Lynceus Laevicaudata Thorp & Rogers 2016 brachyurus) Spinicaudata Genus Thorp & Rogers 2016 Williams 1972, Thorp & Rogers Isopoda Genus 2016 Holsinger 1972, Thorp & Rogers Amphipoda Genus 2016 Gammaridae: Gammarus Species Holsinger 1972 Crustacea monotypic genera: Apocorophium lacustre, Echinogammarus ischnus, Synurella dentata Species (Taphromysis Mysida Thorp & Rogers 2016 louisianae) Crocker & Barr 1968; Jezerinac 1993, 1995; Jezerinac & Thoma 1984; Taylor 2000; Thoma et al. Cambaridae Species 2005; Thoma & Stocker 2009; Crandall & De Grave 2017; Glon et al. 2018 Species (Palaemon Pennak 1989, Palaemonidae kadiakensis) Thorp & Rogers 2016 1 Ohio EPA Macroinvertebrate Taxonomic Level December 2019 Taxon Subtaxon Taxonomic Level Taxonomic Key(ies) Informal grouping of the Arachnida Hydrachnidia Smith 2001 water mites Genus Morse et al. -

Table of Contents 2
Southwest Association of Freshwater Invertebrate Taxonomists (SAFIT) List of Freshwater Macroinvertebrate Taxa from California and Adjacent States including Standard Taxonomic Effort Levels 1 March 2011 Austin Brady Richards and D. Christopher Rogers Table of Contents 2 1.0 Introduction 4 1.1 Acknowledgments 5 2.0 Standard Taxonomic Effort 5 2.1 Rules for Developing a Standard Taxonomic Effort Document 5 2.2 Changes from the Previous Version 6 2.3 The SAFIT Standard Taxonomic List 6 3.0 Methods and Materials 7 3.1 Habitat information 7 3.2 Geographic Scope 7 3.3 Abbreviations used in the STE List 8 3.4 Life Stage Terminology 8 4.0 Rare, Threatened and Endangered Species 8 5.0 Literature Cited 9 Appendix I. The SAFIT Standard Taxonomic Effort List 10 Phylum Silicea 11 Phylum Cnidaria 12 Phylum Platyhelminthes 14 Phylum Nemertea 15 Phylum Nemata 16 Phylum Nematomorpha 17 Phylum Entoprocta 18 Phylum Ectoprocta 19 Phylum Mollusca 20 Phylum Annelida 32 Class Hirudinea Class Branchiobdella Class Polychaeta Class Oligochaeta Phylum Arthropoda Subphylum Chelicerata, Subclass Acari 35 Subphylum Crustacea 47 Subphylum Hexapoda Class Collembola 69 Class Insecta Order Ephemeroptera 71 Order Odonata 95 Order Plecoptera 112 Order Hemiptera 126 Order Megaloptera 139 Order Neuroptera 141 Order Trichoptera 143 Order Lepidoptera 165 2 Order Coleoptera 167 Order Diptera 219 3 1.0 Introduction The Southwest Association of Freshwater Invertebrate Taxonomists (SAFIT) is charged through its charter to develop standardized levels for the taxonomic identification of aquatic macroinvertebrates in support of bioassessment. This document defines the standard levels of taxonomic effort (STE) for bioassessment data compatible with the Surface Water Ambient Monitoring Program (SWAMP) bioassessment protocols (Ode, 2007) or similar procedures. -
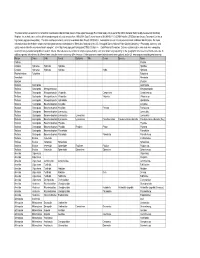
This Table Contains a Taxonomic List of Benthic Invertebrates Collected from Streams in the Upper Mississippi River Basin Study
This table contains a taxonomic list of benthic invertebrates collected from streams in the Upper Mississippi River Basin study unit as part of the USGS National Water Quality Assessemnt (NAWQA) Program. Invertebrates were collected from woody snags in selected streams from 1996-2004. Data Retreival occurred 26-JAN-06 11.10.25 AM from the USGS data warehouse (Taxonomic List Invert http://water.usgs.gov/nawqa/data). The data warehouse currently contains invertebrate data through 09/30/2002. Invertebrate taxa can include provisional and conditional identifications. For more information about invertebrate sample processing and taxonomic standards see, "Methods of analysis by the U.S. Geological Survey National Water Quality Laboratory -- Processing, taxonomy, and quality control of benthic macroinvertebrate samples", at << http://nwql.usgs.gov/Public/pubs/OFR00-212.html >>. Data Retrieval Precaution: Extreme caution must be exercised when comparing taxonomic lists generated using different search criteria. This is because the number of samples represented by each taxa list will vary depending on the geographic criteria selected for the retrievals. In addition, species lists retrieved at different times using the same criteria may differ because: (1) the taxonomic nomenclature (names) were updated, and/or (2) new samples containing new taxa may Phylum Class Order Family Subfamily Tribe Genus Species Taxon Porifera Porifera Cnidaria Hydrozoa Hydroida Hydridae Hydridae Cnidaria Hydrozoa Hydroida Hydridae Hydra Hydra sp. Platyhelminthes Turbellaria Turbellaria Nematoda Nematoda Bryozoa Bryozoa Mollusca Gastropoda Gastropoda Mollusca Gastropoda Mesogastropoda Mesogastropoda Mollusca Gastropoda Mesogastropoda Viviparidae Campeloma Campeloma sp. Mollusca Gastropoda Mesogastropoda Viviparidae Viviparus Viviparus sp. Mollusca Gastropoda Mesogastropoda Hydrobiidae Hydrobiidae Mollusca Gastropoda Basommatophora Ancylidae Ancylidae Mollusca Gastropoda Basommatophora Ancylidae Ferrissia Ferrissia sp. -

THE CHIRONOMIDAE of OTSEGO LAKE with KEYS to the IMMATURE STAGES of the SUBFAMILIES TANYPODINAE and DIAMESINAE (DIPTERA) Joseph
THE CHIRONOMIDAE OF OTSEGO LAKE WITH KEYS TO THE IMMATURE STAGES OF THE SUBFAMILIES TANYPODINAE AND DIAMESINAE (DIPTERA) Joseph P. Fagnani Willard N. Harman BIOLOGICAL FIELD STATION COOPERSTOWN, NEW YORK OCCASIONAL PAPER NO. 20 AUGUST, 1987 BIOLOGY DEPARTMENT STATE UNIVERSITY COLLEGE AT ONEONTA THIS MANUSCRIPT IS NOT A FORMAL PUBLICATION. The information contained herein may not be cited or reproduced without permission of the author or the S.U.N.Y. Oneonta Biology Department ABSTRACT The species of Chironomidae inhabiting Otsego Lake, New York, were studied from 1979 through 1982. This report presents the results of a variety of collecting methods used in a diversity of habitats over a considerable temporal period. The principle emphasis was on sound taxonomy and rearing to associate immatures and adults. Over 4,000 individual rearings have provided the basis for description of general morphological stages that occur during the life cycles of these species. Keys to the larvae and pupae of the 4 subfamilies and 10 tribes of Chironomidae collected in Otsego Lake were compiled. Keys are also presented for the immature stages of 12 Tanypodinae and 2 Diamesinae species found in Otsego Lake. Labeled line drawings of the majority of structures and measurements used to identify the immature stages of most species of chironomids were adapted from the literature. Extensive photomicrographs are presented along with larval and pupal characteristics, taxonomic notes, synonymies, recent literature accounts and collection records for 17 species. These include: Chironominae Paratendipes albimanus (Meigen) (Chironomini); Ortl10cladiinae Psectrocladius (Psectrocladius) simulans (Johannsen) and I!ydrobaenus johannseni (Sublette); Diamesinae - Protanypus ramosus Saether (Protanypodini) and Potthastia longimana Kieffer (Diamesini); and Tanypodinae - Clinotanypus (Clinotanypus) pinguis (Loew) (Coelotanypodini), Tanypu~ (Tanypus) punctipennis Meigen (Tanypodini), Procladius (Psilotanypus) bellus (Loew); Var. -

Wiscoy Creek, 2015
WISCOY CREEK Biological Stream Assessment April 1, 2015 STREAM BIOMONITORING UNIT 425 Jordan Rd, Troy, NY 12180 P: (518) 285-5627 | F: (518) 285-5601 | [email protected] www.dec.ny.gov BIOLOGICAL STREAM ASSESSMENT Wiscoy Creek Wyoming and Allegany Counties, New York Genesee River Basin Survey date: June 25-26, 2014 Report date: April 1, 2015 Alexander J. Smith Elizabeth A. Mosher Mirian Calderon Jeff L. Lojpersberger Diana L. Heitzman Brian T. Duffy Margaret A. Novak Stream Biomonitoring Unit Bureau of Water Assessment and Management Division of Water NYS Department of Environmental Conservation Albany, New York www.dec.ny.gov For additional information regarding this report please contact: Alexander J. Smith, PhD New York State Department of Environmental Conservation Stream Biomonitoring Unit 425 Jordan Road, Troy, NY 12180 [email protected] ph 518-285-5627 fx 518-285-5601 Table of Contents Stream ............................................................................................................................................. 1 River Basin...................................................................................................................................... 1 Reach............................................................................................................................................... 1 Background ..................................................................................................................................... 1 Results and Conclusions ................................................................................................................ -

The Species of the Genus Diamesa (Diptera, Chironomidae) Known to Occur in Italian Alps and Apennines
Zootaxa 4193 (2): 317–331 ISSN 1175-5326 (print edition) http://www.mapress.com/j/zt/ Article ZOOTAXA Copyright © 2016 Magnolia Press ISSN 1175-5334 (online edition) http://doi.org/10.11646/zootaxa.4193.2.7 http://zoobank.org/urn:lsid:zoobank.org:pub:A1A56E2F-3A9A-4A07-B3BB-EE50B669FB50 The species of the genus Diamesa (Diptera, Chironomidae) known to occur in Italian Alps and Apennines MATTEO MONTAGNA1, SANDRA URBANELLI2 & BRUNO ROSSARO3,4 1Dipartimento di Scienze Agrarie e Ambientali—Università degli Studi di Milano, Via Celoria 2, I-20133, Milano, Italy 2Department of Environmental Biology, Università "La Sapienza" of Rome, Italy 3Dipartimento di Scienze per gli Alimenti, la Nutrizione e l’Ambiente—Università degli Studi di Milano, Via Celoria 2, I-20133, Milano, Italy 4Corresponding author. E-mail: [email protected] Abstract Some rare species from Italian Alps, belonging to the genus Diamesa Meigen, 1835 (Diptera, Chironomidae) are here re- described as adult males, because only old, incomplete descriptions are available for these taxa. Terminology of male gen- italia is reviewed, diagnostic features are illustrated in detail, and notes on biology and geographical distribution of the examined species are provided. An identification key to the known adult males is presented. Key words: Diamesinae, Diamesini, taxonomy, aquatic insects, male genitalia Introduction New conservation policies for freshwater ecosystems need to be developed as a priority due to the presence of rare species with high conservation value. Growing threats to their existence require attention, considering their importance in maintaining natural food webs (Moore et al. 2009). Effective conservation strategies require the appropriate knowledge on the morphology of the rare species as well as an update of their distribution and ecology. -

F. Christian Thompson Neal L. Evenhuis and Curtis W. Sabrosky Bibliography of the Family-Group Names of Diptera
F. Christian Thompson Neal L. Evenhuis and Curtis W. Sabrosky Bibliography of the Family-Group Names of Diptera Bibliography Thompson, F. C, Evenhuis, N. L. & Sabrosky, C. W. The following bibliography gives full references to 2,982 works cited in the catalog as well as additional ones cited within the bibliography. A concerted effort was made to examine as many of the cited references as possible in order to ensure accurate citation of authorship, date, title, and pagination. References are listed alphabetically by author and chronologically for multiple articles with the same authorship. In cases where more than one article was published by an author(s) in a particular year, a suffix letter follows the year (letters are listed alphabetically according to publication chronology). Authors' names: Names of authors are cited in the bibliography the same as they are in the text for proper association of literature citations with entries in the catalog. Because of the differing treatments of names, especially those containing articles such as "de," "del," "van," "Le," etc., these names are cross-indexed in the bibliography under the various ways in which they may be treated elsewhere. For Russian and other names in Cyrillic and other non-Latin character sets, we follow the spelling used by the authors themselves. Dates of publication: Dating of these works was obtained through various methods in order to obtain as accurate a date of publication as possible for purposes of priority in nomenclature. Dates found in the original works or by outside evidence are placed in brackets after the literature citation. -

Diamesa Amanoi Sp. П., a New Species of Diamesinae (Diptera, Chironomidae) from Nepal, with Notes on Taxonomy and Distribution of Some Diamesa Meigen
[Med. Entomol. Zool. Vol. 48 No. 1 p. 45-48 1997) Diamesa amanoi sp. п., a new species of Diamesinae (Diptera, Chironomidae) from Nepal, with notes on taxonomy and distribution of some Diamesa Meigen Eugenyi A. MAKARCHENKO1) and Tadashi KoBAYASHI2) 1) Institute of biology and Pedology, Far East Branch of Russian Academy of Sciences, Vladivostok, 690022, Russia 2) Kanagawa University High School, 800 Daimura-cho Midori-ku, Yokohama, 226 Japan (Received: 6 November 1996; Accepted: 20 January 1997) Key words: Chironomidae, Diamesinae, Diamesa, new species, Nepal, taxonomy Abstract: A new species, Diamesa amanoi sp. п., from Mera La (alt. 5,050 m), Nepal is described. The present new species is most closely related to D. subletti Makarchenko from North America. Diamesa plumicornis Tokunaga which had been regarded as Japanese endemic species is newly recorded from South Korea, and D. pankratovae Makarchenko et Bulgakov is recognized as a new synonym of D. filicauda Tokunaga. number: CBM-ZI 72725). Paratypes are INTRODUCTION deposited in Institute of Biology and Pe- dology, Far East Branch of Russian Acad- Up to the present time the Nepalese sub- emy of Sciences, Vladivostok, Russia. family Diamesinae are represented by the following 15 species: Boreoheptagyia rotun- Diamesa amanoi Makarchenko da Serra-Tosio, B. sp., Diamesa aberrata et Kobayashi, sp. n. Lundbeck, D. barraudi Pagast, D. khum- (Figs. 1-5) bugelida Saether et Willassen, D. koshimai Saether et Willassen, D. loeffleri Reiss, D. Male. General color dark brown. Body planistyla Reiss., D. praedpua Saether et length 3.4-3.6 mm, body length/wing Willassen, D. yalavia Saether et Willassen, length 0.9-1.0. -

Chironomidae of the Southeastern United States: a Checklist of Species and Notes on Biology, Distribution, and Habitat
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln US Fish & Wildlife Publications US Fish & Wildlife Service 1990 Chironomidae of the Southeastern United States: A Checklist of Species and Notes on Biology, Distribution, and Habitat Patrick L. Hudson U.S. Fish and Wildlife Service David R. Lenat North Carolina Department of Natural Resources Broughton A. Caldwell David Smith U.S. Evironmental Protection Agency Follow this and additional works at: https://digitalcommons.unl.edu/usfwspubs Part of the Aquaculture and Fisheries Commons Hudson, Patrick L.; Lenat, David R.; Caldwell, Broughton A.; and Smith, David, "Chironomidae of the Southeastern United States: A Checklist of Species and Notes on Biology, Distribution, and Habitat" (1990). US Fish & Wildlife Publications. 173. https://digitalcommons.unl.edu/usfwspubs/173 This Article is brought to you for free and open access by the US Fish & Wildlife Service at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in US Fish & Wildlife Publications by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Fish and Wildlife Research 7 Chironomidae of the Southeastern United States: A Checklist of Species and Notes on Biology, Distribution, and Habitat NWRC Library I7 49.99:- -------------UNITED STATES DEPARTMENT OF THE INTERIOR FISH AND WILDLIFE SERVICE Fish and Wildlife Research This series comprises scientific and technical reports based on original scholarly research, interpretive reviews, or theoretical presentations. Publications in this series generally relate to fish or wildlife and their ecology. The Service distributes these publications to natural resource agencies, libraries and bibliographic collection facilities, scientists, and resource managers. Copies of this publication may be obtained from the Publications Unit, U.S. -

Refinement of the Basin-Wide Index of Biotic Integrity for Non-Tidal Streams and Wadeable Rivers in the Chesapeake Bay Watershed
Refinement of the Basin-Wide Index of Biotic Integrity for Non-Tidal Streams and Wadeable Rivers in the Chesapeake Bay Watershed APPENDICES Appendix A: Taxonomic Classification Appendix B: Taxonomic Attributes Appendix C: Taxonomic Standardization Appendix D: Rarefaction Appendix E: Biological Metric Descriptions Appendix F: Abiotic Parameters for Evaluating Stream Environment Appendix G: Stream Classification Appendix H: HUC12 Watershed Characteristics in Bioregions Appendix I: Index Methodologies Appendix J: Scoring Methodologies Appendix K: Index Performance, Accuracy, and Precision Appendix L: Narrative Ratings and Maps of Index Scores Appendix M: Potential Biases in the Regional Index Ratings Appendix Citations Appendix A: Taxonomic Classification All taxa reported in Chessie BIBI database were assigned the appropriate Phylum, Subphylum, Class, Subclass, Order, Suborder, Family, Subfamily, Tribe, and Genus when applicable. A portion of the taxa reported were reported under an invalid name according to the ITIS database. These taxa were subsequently changed to the taxonomic name deemed valid by ITIS. Table A-1. The taxonomic hierarchy of stream macroinvertebrate taxa included in the Chesapeake Bay non-tidal database.