Mir-153 Regulates Amelogenesis by Targeting Endocytotic And
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Anti-Rab11 Antibody (ARG41900)
Product datasheet [email protected] ARG41900 Package: 100 μg anti-Rab11 antibody Store at: -20°C Summary Product Description Goat Polyclonal antibody recognizes Rab11 Tested Reactivity Hu, Ms, Rat, Dog, Mk Tested Application IHC-Fr, IHC-P, WB Host Goat Clonality Polyclonal Isotype IgG Target Name Rab11 Antigen Species Mouse Immunogen Purified recombinant peptides within aa. 110 to the C-terminus of Mouse Rab11a, Rab11b and Rab11c (Rab25). Conjugation Un-conjugated Alternate Names RAB11A: Rab-11; Ras-related protein Rab-11A; YL8 RAB11B: GTP-binding protein YPT3; H-YPT3; Ras-related protein Rab-11B RAB25: RAB11C; CATX-8; Ras-related protein Rab-25 Application Instructions Application table Application Dilution IHC-Fr 1:100 - 1:400 IHC-P 1:100 - 1:400 WB 1:250 - 1:2000 Application Note IHC-P: Antigen Retrieval: Heat mediation was recommended. * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. Positive Control Hepa cell lysate Calculated Mw 24 kDa Observed Size ~ 26 kDa Properties Form Liquid Purification Affinity purification with immunogen. Buffer PBS, 0.05% Sodium azide and 20% Glycerol. Preservative 0.05% Sodium azide www.arigobio.com 1/3 Stabilizer 20% Glycerol Concentration 3 mg/ml Storage instruction For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C. Storage in frost free freezers is not recommended. Avoid repeated freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use. Note For laboratory research only, not for drug, diagnostic or other use. -

Mouse Rab11fip2 Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Rab11fip2 Knockout Project (CRISPR/Cas9) Objective: To create a Rab11fip2 knockout Mouse model (C57BL/6J) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Rab11fip2 gene (NCBI Reference Sequence: NM_001164367 ; Ensembl: ENSMUSG00000040022 ) is located on Mouse chromosome 19. 5 exons are identified, with the ATG start codon in exon 2 and the TAG stop codon in exon 5 (Transcript: ENSMUST00000171986). Exon 2 will be selected as target site. Cas9 and gRNA will be co-injected into fertilized eggs for KO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Exon 2 starts from the coding region. Exon 2 covers 33.33% of the coding region. The size of effective KO region: ~443 bp. The KO region does not have any other known gene. Page 1 of 8 https://www.alphaknockout.com Overview of the Targeting Strategy Wildtype allele gRNA region 5' gRNA region 3' 1 2 5 Legends Exon of mouse Rab11fip2 Knockout region Page 2 of 8 https://www.alphaknockout.com Overview of the Dot Plot (up) Window size: 15 bp Forward Reverse Complement Sequence 12 Note: The 2000 bp section upstream of Exon 2 is aligned with itself to determine if there are tandem repeats. No significant tandem repeat is found in the dot plot matrix. So this region is suitable for PCR screening or sequencing analysis. Overview of the Dot Plot (down) Window size: 15 bp Forward Reverse Complement Sequence 12 Note: The 913 bp section downstream of Exon 2 is aligned with itself to determine if there are tandem repeats. -

An Advance About the Genetic Causes of Epilepsy
E3S Web of Conferences 271, 03068 (2021) https://doi.org/10.1051/e3sconf/202127103068 ICEPE 2021 An advance about the genetic causes of epilepsy Yu Sun1, a, *, †, Licheng Lu2, b, *, †, Lanxin Li3, c, *, †, Jingbo Wang4, d, *, † 1The School of Molecular and Cellular Biology, University of Illinois at Urbana-Champaign, Urbana, IL 61801-3633, US 2High School Affiliated to Shanghai Jiao Tong University, Shanghai, 200441, China 3Applied Biology program, University of British Columbia, Vancouver, V6r3b1, Canada 4School of Chemical Machinery and Safety, Dalian University of Technology, Dalian, 116023, China †These authors contributed equally. Abstract: Human hereditary epilepsy has been found related to ion channel mutations in voltage-gated channels (Na+, K+, Ca2+, Cl-), ligand gated channels (GABA receptors), and G-protein coupled receptors, such as Mass1. In addition, some transmembrane proteins or receptor genes, including PRRT2 and nAChR, and glucose transporter genes, such as GLUT1 and SLC2A1, are also about the onset of epilepsy. The discovery of these genetic defects has contributed greatly to our understanding of the pathology of epilepsy. This review focuses on introducing and summarizing epilepsy-associated genes and related findings in recent decades, pointing out related mutant genes that need to be further studied in the future. 1 Introduction Epilepsy is a neurological disorder characterized by 2 Malfunction of Ion channel epileptic seizures caused by abnormal brain activity. 1 in Functional variation in voltage or ligand-gated ion 100 (50 million people) people are affected by symptoms channel mutations is a major cause of idiopathic epilepsy, of this disorder worldwide, with men, young children, and especially in rare genetic forms. -

Downloaded the “Top Edge” Version
bioRxiv preprint doi: https://doi.org/10.1101/855338; this version posted December 6, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 Drosophila models of pathogenic copy-number variant genes show global and 2 non-neuronal defects during development 3 Short title: Non-neuronal defects of fly homologs of CNV genes 4 Tanzeen Yusuff1,4, Matthew Jensen1,4, Sneha Yennawar1,4, Lucilla Pizzo1, Siddharth 5 Karthikeyan1, Dagny J. Gould1, Avik Sarker1, Yurika Matsui1,2, Janani Iyer1, Zhi-Chun Lai1,2, 6 and Santhosh Girirajan1,3* 7 8 1. Department of Biochemistry and Molecular Biology, Pennsylvania State University, 9 University Park, PA 16802 10 2. Department of Biology, Pennsylvania State University, University Park, PA 16802 11 3. Department of Anthropology, Pennsylvania State University, University Park, PA 16802 12 4 contributed equally to work 13 14 *Correspondence: 15 Santhosh Girirajan, MBBS, PhD 16 205A Life Sciences Building 17 Pennsylvania State University 18 University Park, PA 16802 19 E-mail: [email protected] 20 Phone: 814-865-0674 21 1 bioRxiv preprint doi: https://doi.org/10.1101/855338; this version posted December 6, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 22 ABSTRACT 23 While rare pathogenic copy-number variants (CNVs) are associated with both neuronal and non- 24 neuronal phenotypes, functional studies evaluating these regions have focused on the molecular 25 basis of neuronal defects. -

Mir-15A and Mir-16-1 Cluster Functions in Human Leukemia
MiR-15a and miR-16-1 cluster functions in human leukemia George A. Calin*†‡, Amelia Cimmino*§, Muller Fabbri*, Manuela Ferracin¶, Sylwia E. Wojcik*, Masayoshi Shimizu*§, Cristian Taccioli*, Nicola Zanesi*, Ramiro Garzon*, Rami I. Aqeilan*, Hansjuerg Alder*, Stefano Volinia*ʈ, Laura Rassenti**, Xiuping Liu*, Chang-gong Liu*, Thomas J. Kipps**, Massimo Negrini¶, and Carlo M. Croce*†† *Human Cancer Genetics Program and †Department of Molecular Virology, Immunology, and Medical Genetics, Ohio State University, Columbus, OH 43210; §Department of Biochemistry and Biophysics ‘‘F. Cedrangolo,’’ Medical School, Second University of Naples, 80138 Naples, Italy; ¶Department of Experimental and Diagnostic Medicine, Interdepartment Center for Cancer Research and ʈMorphology and Embryology Department, University of Ferrara, 44100 Ferrara, Italy; and **Department of Medicine, University of California at San Diego, La Jolla, CA 92093 Contributed by Carlo M. Croce, January 16, 2008 (sent for review December 6, 2007) MicroRNAs (miRNAs) are short noncoding RNAs regulating gene megakaryocytic leukemia (22). These data support a role for expression that play roles in human diseases, including cancer. Each miR-15a and miR-16-1 as tumor-suppressor genes (TSGs) in CLLs miRNA is predicted to regulate hundreds of transcripts, but only few and perhaps in other malignancies in which these genes are lost or have experimental validation. In chronic lymphocytic leukemia (CLL), down-regulated. the most common adult human leukemia, miR-15a and miR-16-1 are Here, to investigate the mechanism of action of miR-15a and lost or down-regulated in the majority of cases. After our previous miR-16-1 as tumor suppressors in leukemias, we analyzed the work indicating a tumor suppressor function of miR-15a/16-1 by effects of miR-15a and miR-16-1 on transcriptome and proteome in targeting the BCL2 oncogene, here, we produced a high-throughput MEG-01 leukemic cells. -

Ion Channels
UC Davis UC Davis Previously Published Works Title THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Ion channels. Permalink https://escholarship.org/uc/item/1442g5hg Journal British journal of pharmacology, 176 Suppl 1(S1) ISSN 0007-1188 Authors Alexander, Stephen PH Mathie, Alistair Peters, John A et al. Publication Date 2019-12-01 DOI 10.1111/bph.14749 License https://creativecommons.org/licenses/by/4.0/ 4.0 Peer reviewed eScholarship.org Powered by the California Digital Library University of California S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2019/20: Ion channels. British Journal of Pharmacology (2019) 176, S142–S228 THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Ion channels Stephen PH Alexander1 , Alistair Mathie2 ,JohnAPeters3 , Emma L Veale2 , Jörg Striessnig4 , Eamonn Kelly5, Jane F Armstrong6 , Elena Faccenda6 ,SimonDHarding6 ,AdamJPawson6 , Joanna L Sharman6 , Christopher Southan6 , Jamie A Davies6 and CGTP Collaborators 1School of Life Sciences, University of Nottingham Medical School, Nottingham, NG7 2UH, UK 2Medway School of Pharmacy, The Universities of Greenwich and Kent at Medway, Anson Building, Central Avenue, Chatham Maritime, Chatham, Kent, ME4 4TB, UK 3Neuroscience Division, Medical Education Institute, Ninewells Hospital and Medical School, University of Dundee, Dundee, DD1 9SY, UK 4Pharmacology and Toxicology, Institute of Pharmacy, University of Innsbruck, A-6020 Innsbruck, Austria 5School of Physiology, Pharmacology and Neuroscience, University of Bristol, Bristol, BS8 1TD, UK 6Centre for Discovery Brain Science, University of Edinburgh, Edinburgh, EH8 9XD, UK Abstract The Concise Guide to PHARMACOLOGY 2019/20 is the fourth in this series of biennial publications. The Concise Guide provides concise overviews of the key properties of nearly 1800 human drug targets with an emphasis on selective pharmacology (where available), plus links to the open access knowledgebase source of drug targets and their ligands (www.guidetopharmacology.org), which provides more detailed views of target and ligand properties. -

Pflugers Final
CORE Metadata, citation and similar papers at core.ac.uk Provided by Serveur académique lausannois A comprehensive analysis of gene expression profiles in distal parts of the mouse renal tubule. Sylvain Pradervand2, Annie Mercier Zuber1, Gabriel Centeno1, Olivier Bonny1,3,4 and Dmitri Firsov1,4 1 - Department of Pharmacology and Toxicology, University of Lausanne, 1005 Lausanne, Switzerland 2 - DNA Array Facility, University of Lausanne, 1015 Lausanne, Switzerland 3 - Service of Nephrology, Lausanne University Hospital, 1005 Lausanne, Switzerland 4 – these two authors have equally contributed to the study to whom correspondence should be addressed: Dmitri FIRSOV Department of Pharmacology and Toxicology, University of Lausanne, 27 rue du Bugnon, 1005 Lausanne, Switzerland Phone: ++ 41-216925406 Fax: ++ 41-216925355 e-mail: [email protected] and Olivier BONNY Department of Pharmacology and Toxicology, University of Lausanne, 27 rue du Bugnon, 1005 Lausanne, Switzerland Phone: ++ 41-216925417 Fax: ++ 41-216925355 e-mail: [email protected] 1 Abstract The distal parts of the renal tubule play a critical role in maintaining homeostasis of extracellular fluids. In this review, we present an in-depth analysis of microarray-based gene expression profiles available for microdissected mouse distal nephron segments, i.e., the distal convoluted tubule (DCT) and the connecting tubule (CNT), and for the cortical portion of the collecting duct (CCD) (Zuber et al., 2009). Classification of expressed transcripts in 14 major functional gene categories demonstrated that all principal proteins involved in maintaining of salt and water balance are represented by highly abundant transcripts. However, a significant number of transcripts belonging, for instance, to categories of G protein-coupled receptors (GPCR) or serine-threonine kinases exhibit high expression levels but remain unassigned to a specific renal function. -
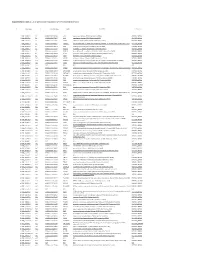
Supplemetary Table 2. List of Genes Down-Regulated in LPAR6 Knocked Down Cells
Supplemetary Table 2. List of genes down-regulated in LPAR6 knocked down cells g# initial alias c# converted alias name description namespace 1 NM_002317.5 1.1 ENSG00000113083 LOX lysyl oxidase [Source:HGNC Symbol;Acc:6664] REFSEQ_MRNA 2 NM_006183.4 2.1 ENSG00000133636 NTS neurotensin [Source:HGNC Symbol;Acc:8038] REFSEQ_MRNA 3 NM_005213.3 3.1 ENSG00000121552 CSTA cystatin A (stefin A) [Source:HGNC Symbol;Acc:2481] REFSEQ_MRNA 4 NM_007231.3 4.1 ENSG00000087916 SLC6A14 solute carrier family 6 (amino acid transporter), member 14 [Source:HGNC Symbol;Acc:11047] REFSEQ_MRNA 5 NM_001873.2 5.1 ENSG00000109472 CPE carboxypeptidase E [Source:HGNC Symbol;Acc:2303] REFSEQ_MRNA 6 NM_019856.1 6.1 ENSG00000101605 MYOM1 myomesin 1, 185kDa [Source:HGNC Symbol;Acc:7613] REFSEQ_MRNA 7 NM_032590.4 7.1 ENSG00000089094 KDM2B lysine (K)-specific demethylase 2B [Source:HGNC Symbol;Acc:13610] REFSEQ_MRNA 8 NM_001901.2 8.1 ENSG00000118523 CTGF connective tissue growth factor [Source:HGNC Symbol;Acc:2500] REFSEQ_MRNA 9 NM_007183.2 9.1 ENSG00000184363 PKP3 plakophilin 3 [Source:HGNC Symbol;Acc:9025] REFSEQ_MRNA 10 NM_182965.2 10.1 ENSG00000176170 SPHK1 sphingosine kinase 1 [Source:HGNC Symbol;Acc:11240] REFSEQ_MRNA 11 NM_152423.4 11.1 ENSG00000157502 MUM1L1 melanoma associated antigen (mutated) 1-like 1 [Source:HGNC Symbol;Acc:26583] REFSEQ_MRNA 12 NM_002923.3 12.1 ENSG00000116741 RGS2 regulator of G-protein signaling 2, 24kDa [Source:HGNC Symbol;Acc:9998] REFSEQ_MRNA 13 NR_003038.2 13.1 N/A N/A N/A N/A 14 NM_080862.1 14.1 ENSG00000175093 SPSB4 splA/ryanodine receptor -

Gene List of the Targeted NGS MCD and CCA Gene Panel AKT3,ALX1
Gene List of the targeted NGS MCD and CCA gene panel AKT3,ALX1,ALX3,ALX4,AMPD2,ARFGEF2,ARID1B,ARX,ASPM,ATR,ATRX,B3GALTL,BRPF1,c12orf57,C6orf70,CASK,CCND2,CDK5RAP2,CDON,C ENPJ,CEP170,CHMP1A,COL4A1,CREBBP,CYP11A1,DCHS1,DCLK1,DCX,DHCR24,DHCR7,DIS3L2,DISC1,DISP1,DLL1,DMRTA2,DYNC1H1,DYRK1 A,EARS2,EFNB1,EMX1,EOMES,EP300,ERBB4,ERMARD,EXOSC3,FAM36A,FGF8,FGFR1,FGFR2,FLNA,FOXC1,FOXG1,FOXH1,FZD10,GLI2,GLI3,GP R56,GPSM2,HCCS,HESX1,HNRNPU,IGBP1,IGFBP1,ISPD,ITPA,KAL1,KAT6B,KATNB1,KIAA1279,KIF14,KIF1A,KIF1B,KIF21A,KIF2A,KIF5C,KIF7,L1 CAM,LAMB1,LAMC3,LRP2,MCPH1,MED12,MID1,NDE1,NFIB,NPC1,NR2F1,NSD1,NTRK1,NTRK3,OCEL1,OPA1,OTX2,PAFAH1B1,PAX6,PEX1,PHF1 0,PIK3R2,POLR3A,POLR3B,POMT1,POMT2,PTCH1,PTPRS,PYCR1,RAB3GAP1,RARS2,RELN,RFX3,ROBO1,ROBO3,RPS6KA3,RTTN,SATB2,SEPSEC S,SHH,SIX3,SLC12A6,SOX2,SPOCK1,SRPX2,TBCD,TBCE,TCF4,TDGF1,TEAD1,THBS2,TMEM5,TSC1,TSC2,TSEN15,TSEN2,TSEN34,TSEN54,TUBA1 A,TUBA8,TUBB,TUBB2A,TUBB2B,TUBB3,TUBB4A,TUBG1,VAX1,VRK1,WDR47,WDR62,ZBTB18,ZEB2,ZIC2. Gene List of the targeted NGS epilepsy gene panel AARS, ADGRV1, ADRA2B, ADSL, ALDH4A1, ALDH7A1, ALG13, ALPL, ARHGEF15, ARHGEF9, ARX, ASAH1, ATP1A2, ATP1A3, BRD2, CACNA1A, CACNA1H, CACNA2D2, CACNB4, CBL, CDKL5, CERS1, CHD2, CHRNA2, CHRNA4, CHRNB2, CLCN2, CLCN4, CLN8, CLTC, CNKSR2, CNTNAP2, CPA6, CPLX1, CSNK1G1, CSNK2B, CTNND2, DEPDC5, DHDDS, DNM1, DOCK7, DYNC1H1, EEF1A2, EFHC1, EIF2S3, EMC1, EPM2A, FASN, FLNA, FOXG1, GABBR2, GABRA1, GABRA2, GABRA3, GABRB2, GABRB3, GABRD, GABRG2, GAL, GNAO1, GOSR2, GRIA1, GRIN1, GRIN2A, GRIN2B, HCN1, HCN4, HDAC4, HNRNPU, IDH3A, IQSEC2, JRK, KCNA1, KCNA2, KCNB1, -

CENTOGENE's Severe and Early Onset Disorder Gene List
CENTOGENE’s severe and early onset disorder gene list USED IN PRENATAL WES ANALYSIS AND IDENTIFICATION OF “PATHOGENIC” AND “LIKELY PATHOGENIC” CENTOMD® VARIANTS IN NGS PRODUCTS The following gene list shows all genes assessed in prenatal WES tests or analysed for P/LP CentoMD® variants in NGS products after April 1st, 2020. For searching a single gene coverage, just use the search on www.centoportal.com AAAS, AARS1, AARS2, ABAT, ABCA12, ABCA3, ABCB11, ABCB4, ABCB7, ABCC6, ABCC8, ABCC9, ABCD1, ABCD4, ABHD12, ABHD5, ACACA, ACAD9, ACADM, ACADS, ACADVL, ACAN, ACAT1, ACE, ACO2, ACOX1, ACP5, ACSL4, ACTA1, ACTA2, ACTB, ACTG1, ACTL6B, ACTN2, ACVR2B, ACVRL1, ACY1, ADA, ADAM17, ADAMTS2, ADAMTSL2, ADAR, ADARB1, ADAT3, ADCY5, ADGRG1, ADGRG6, ADGRV1, ADK, ADNP, ADPRHL2, ADSL, AFF2, AFG3L2, AGA, AGK, AGL, AGPAT2, AGPS, AGRN, AGT, AGTPBP1, AGTR1, AGXT, AHCY, AHDC1, AHI1, AIFM1, AIMP1, AIPL1, AIRE, AK2, AKR1D1, AKT1, AKT2, AKT3, ALAD, ALDH18A1, ALDH1A3, ALDH3A2, ALDH4A1, ALDH5A1, ALDH6A1, ALDH7A1, ALDOA, ALDOB, ALG1, ALG11, ALG12, ALG13, ALG14, ALG2, ALG3, ALG6, ALG8, ALG9, ALMS1, ALOX12B, ALPL, ALS2, ALX3, ALX4, AMACR, AMER1, AMN, AMPD1, AMPD2, AMT, ANK2, ANK3, ANKH, ANKRD11, ANKS6, ANO10, ANO5, ANOS1, ANTXR1, ANTXR2, AP1B1, AP1S1, AP1S2, AP3B1, AP3B2, AP4B1, AP4E1, AP4M1, AP4S1, APC2, APTX, AR, ARCN1, ARFGEF2, ARG1, ARHGAP31, ARHGDIA, ARHGEF9, ARID1A, ARID1B, ARID2, ARL13B, ARL3, ARL6, ARL6IP1, ARMC4, ARMC9, ARSA, ARSB, ARSL, ARV1, ARX, ASAH1, ASCC1, ASH1L, ASL, ASNS, ASPA, ASPH, ASPM, ASS1, ASXL1, ASXL2, ASXL3, ATAD3A, ATCAY, ATIC, ATL1, ATM, ATOH7, -

Nucleotide Bound to Rab11a Controls Localization in Rod Cells but Not Interaction with Rhodopsin
14854 • The Journal of Neuroscience, November 5, 2014 • 34(45):14854–14863 Cellular/Molecular Nucleotide Bound to rab11a Controls Localization in Rod Cells But Not Interaction with Rhodopsin Nicholas J. Reish,1,2 Evan R. Boitet,1,3 Katie L. Bales,1,3 and XAlecia K. Gross1,2,3 1Evelyn F. McKnight Brain Institute, 2Department of Neurobiology, and 3Department of Vision Sciences, School of Optometry, University of Alabama at Birmingham, Birmingham, Alabama 35294 Precise vectorial transport of rhodopsin is essential for rod photoreceptor health and function. Mutations that truncate or extend the C terminus of rhodopsin disrupt this transport, and lead to retinal degeneration and blindness in human patients and in mouse models. Here we show that such mutations disrupt the binding of rhodopsin to the small GTPase rab11a. The rhodopsin–rab11a interaction is a direct binding interaction that does not depend on the nucleotide binding state of rab11a. Expression of EGFP-rab11a fusion proteins in Xenopus laevis photoreceptors revealed that the nucleotide binding status of rab11a affects its subcellular localization, with GTP-locked mutants concentrated in the inner segment and GDP-locked mutants concentrated in the outer segment. shRNA-mediated knockdown of rab11a in rods led to shortened outer segments and retinal degeneration. Together, our results show the critical importance of direct rhodopsin–rab11a interactions for the formation and maintenance of vertebrate photoreceptors. Key words: protein interactions; protein trafficking; rab11a; rhodopsin Introduction Golgi and the distal inner segment (IS) in lipid vesicles (Deretic Rod photoreceptors sense light using a specialized organelle and Papermaster, 1991) and may traffic into the outer segment known as the outer segment, a unique structure consisting of a (OS) in vesicular form (Sung and Chuang, 2010) or through primary cilium and a stack of closely spaced membranous disks intraflagellar transport (Bhowmick et al., 2009), or by mecha- that are contained within a plasma membrane. -

Gene Trapping Identifies Chloride Channel 4 As a Novel Inducer of Colon Cancer Cell Migration, Invasion and Metastases
British Journal of Cancer (2010) 102, 774 – 782 & 2010 Cancer Research UK All rights reserved 0007 – 0920/10 $32.00 www.bjcancer.com Gene trapping identifies chloride channel 4 as a novel inducer of colon cancer cell migration, invasion and metastases 1 1 2 3 4 *,1 T Ishiguro , H Avila , S-Y Lin , T Nakamura , M Yamamoto and DD Boyd 1 2 Cancer Biology Department, MD Anderson Cancer Center, Houston, TX, USA; Systems Biology Department, MD Anderson Cancer Center, Houston, 3 4 TX, USA; Surgical Oncology Department, Hokkaido University Graduate School of Medicine, Sapporo, Japan; Second Department of Surgery, Hamamatsu University School of Medicine, Hamanatsu, Japan BACKGROUND: To date, there are few reports on gene products contributing to colon cancer progression. METHODS: We used a gene trap comprised of an enhanced retroviral mutagen (ERM) cassette that includes a tetracycline-responsive promoter upstream of a haemagglutinin (HA) tag and a splice donor site. Integration of the ERM within an endogenous gene yields a tetracycline-regulated HA-tagged transcript. We transduced RKO colon cancer cells expressing a tetracycline trans-activator-off with the ERM-encoding retrovirus and screened for enhanced migration. RESULTS: One clone showed fivefold enhanced migration with tetracycline withdrawal. Rapid amplification of cDNA ends identified the trapped gene as the chloride channel 4 (CLCN4) exchanger. Stable expression of a CLCN4 cDNA enhanced motility, whereas cells knocked down or null for this transcript showed reduced migration/invasion. CLCN4-overexpressing RKO colon cancer cells were þ more resistant than controls to proton load-induced cytotoxicity, consistent with the H -extruding function of this antiporter.