Clcn4-2 Genomic Structure Differs Between the X Locus in Mus Spretus and the Autosomal Locus in Mus Musculus: at Motif Enrichment on the X
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

An Advance About the Genetic Causes of Epilepsy
E3S Web of Conferences 271, 03068 (2021) https://doi.org/10.1051/e3sconf/202127103068 ICEPE 2021 An advance about the genetic causes of epilepsy Yu Sun1, a, *, †, Licheng Lu2, b, *, †, Lanxin Li3, c, *, †, Jingbo Wang4, d, *, † 1The School of Molecular and Cellular Biology, University of Illinois at Urbana-Champaign, Urbana, IL 61801-3633, US 2High School Affiliated to Shanghai Jiao Tong University, Shanghai, 200441, China 3Applied Biology program, University of British Columbia, Vancouver, V6r3b1, Canada 4School of Chemical Machinery and Safety, Dalian University of Technology, Dalian, 116023, China †These authors contributed equally. Abstract: Human hereditary epilepsy has been found related to ion channel mutations in voltage-gated channels (Na+, K+, Ca2+, Cl-), ligand gated channels (GABA receptors), and G-protein coupled receptors, such as Mass1. In addition, some transmembrane proteins or receptor genes, including PRRT2 and nAChR, and glucose transporter genes, such as GLUT1 and SLC2A1, are also about the onset of epilepsy. The discovery of these genetic defects has contributed greatly to our understanding of the pathology of epilepsy. This review focuses on introducing and summarizing epilepsy-associated genes and related findings in recent decades, pointing out related mutant genes that need to be further studied in the future. 1 Introduction Epilepsy is a neurological disorder characterized by 2 Malfunction of Ion channel epileptic seizures caused by abnormal brain activity. 1 in Functional variation in voltage or ligand-gated ion 100 (50 million people) people are affected by symptoms channel mutations is a major cause of idiopathic epilepsy, of this disorder worldwide, with men, young children, and especially in rare genetic forms. -

Ion Channels
UC Davis UC Davis Previously Published Works Title THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Ion channels. Permalink https://escholarship.org/uc/item/1442g5hg Journal British journal of pharmacology, 176 Suppl 1(S1) ISSN 0007-1188 Authors Alexander, Stephen PH Mathie, Alistair Peters, John A et al. Publication Date 2019-12-01 DOI 10.1111/bph.14749 License https://creativecommons.org/licenses/by/4.0/ 4.0 Peer reviewed eScholarship.org Powered by the California Digital Library University of California S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2019/20: Ion channels. British Journal of Pharmacology (2019) 176, S142–S228 THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Ion channels Stephen PH Alexander1 , Alistair Mathie2 ,JohnAPeters3 , Emma L Veale2 , Jörg Striessnig4 , Eamonn Kelly5, Jane F Armstrong6 , Elena Faccenda6 ,SimonDHarding6 ,AdamJPawson6 , Joanna L Sharman6 , Christopher Southan6 , Jamie A Davies6 and CGTP Collaborators 1School of Life Sciences, University of Nottingham Medical School, Nottingham, NG7 2UH, UK 2Medway School of Pharmacy, The Universities of Greenwich and Kent at Medway, Anson Building, Central Avenue, Chatham Maritime, Chatham, Kent, ME4 4TB, UK 3Neuroscience Division, Medical Education Institute, Ninewells Hospital and Medical School, University of Dundee, Dundee, DD1 9SY, UK 4Pharmacology and Toxicology, Institute of Pharmacy, University of Innsbruck, A-6020 Innsbruck, Austria 5School of Physiology, Pharmacology and Neuroscience, University of Bristol, Bristol, BS8 1TD, UK 6Centre for Discovery Brain Science, University of Edinburgh, Edinburgh, EH8 9XD, UK Abstract The Concise Guide to PHARMACOLOGY 2019/20 is the fourth in this series of biennial publications. The Concise Guide provides concise overviews of the key properties of nearly 1800 human drug targets with an emphasis on selective pharmacology (where available), plus links to the open access knowledgebase source of drug targets and their ligands (www.guidetopharmacology.org), which provides more detailed views of target and ligand properties. -

Pflugers Final
CORE Metadata, citation and similar papers at core.ac.uk Provided by Serveur académique lausannois A comprehensive analysis of gene expression profiles in distal parts of the mouse renal tubule. Sylvain Pradervand2, Annie Mercier Zuber1, Gabriel Centeno1, Olivier Bonny1,3,4 and Dmitri Firsov1,4 1 - Department of Pharmacology and Toxicology, University of Lausanne, 1005 Lausanne, Switzerland 2 - DNA Array Facility, University of Lausanne, 1015 Lausanne, Switzerland 3 - Service of Nephrology, Lausanne University Hospital, 1005 Lausanne, Switzerland 4 – these two authors have equally contributed to the study to whom correspondence should be addressed: Dmitri FIRSOV Department of Pharmacology and Toxicology, University of Lausanne, 27 rue du Bugnon, 1005 Lausanne, Switzerland Phone: ++ 41-216925406 Fax: ++ 41-216925355 e-mail: [email protected] and Olivier BONNY Department of Pharmacology and Toxicology, University of Lausanne, 27 rue du Bugnon, 1005 Lausanne, Switzerland Phone: ++ 41-216925417 Fax: ++ 41-216925355 e-mail: [email protected] 1 Abstract The distal parts of the renal tubule play a critical role in maintaining homeostasis of extracellular fluids. In this review, we present an in-depth analysis of microarray-based gene expression profiles available for microdissected mouse distal nephron segments, i.e., the distal convoluted tubule (DCT) and the connecting tubule (CNT), and for the cortical portion of the collecting duct (CCD) (Zuber et al., 2009). Classification of expressed transcripts in 14 major functional gene categories demonstrated that all principal proteins involved in maintaining of salt and water balance are represented by highly abundant transcripts. However, a significant number of transcripts belonging, for instance, to categories of G protein-coupled receptors (GPCR) or serine-threonine kinases exhibit high expression levels but remain unassigned to a specific renal function. -
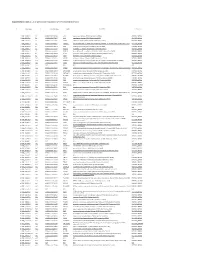
Supplemetary Table 2. List of Genes Down-Regulated in LPAR6 Knocked Down Cells
Supplemetary Table 2. List of genes down-regulated in LPAR6 knocked down cells g# initial alias c# converted alias name description namespace 1 NM_002317.5 1.1 ENSG00000113083 LOX lysyl oxidase [Source:HGNC Symbol;Acc:6664] REFSEQ_MRNA 2 NM_006183.4 2.1 ENSG00000133636 NTS neurotensin [Source:HGNC Symbol;Acc:8038] REFSEQ_MRNA 3 NM_005213.3 3.1 ENSG00000121552 CSTA cystatin A (stefin A) [Source:HGNC Symbol;Acc:2481] REFSEQ_MRNA 4 NM_007231.3 4.1 ENSG00000087916 SLC6A14 solute carrier family 6 (amino acid transporter), member 14 [Source:HGNC Symbol;Acc:11047] REFSEQ_MRNA 5 NM_001873.2 5.1 ENSG00000109472 CPE carboxypeptidase E [Source:HGNC Symbol;Acc:2303] REFSEQ_MRNA 6 NM_019856.1 6.1 ENSG00000101605 MYOM1 myomesin 1, 185kDa [Source:HGNC Symbol;Acc:7613] REFSEQ_MRNA 7 NM_032590.4 7.1 ENSG00000089094 KDM2B lysine (K)-specific demethylase 2B [Source:HGNC Symbol;Acc:13610] REFSEQ_MRNA 8 NM_001901.2 8.1 ENSG00000118523 CTGF connective tissue growth factor [Source:HGNC Symbol;Acc:2500] REFSEQ_MRNA 9 NM_007183.2 9.1 ENSG00000184363 PKP3 plakophilin 3 [Source:HGNC Symbol;Acc:9025] REFSEQ_MRNA 10 NM_182965.2 10.1 ENSG00000176170 SPHK1 sphingosine kinase 1 [Source:HGNC Symbol;Acc:11240] REFSEQ_MRNA 11 NM_152423.4 11.1 ENSG00000157502 MUM1L1 melanoma associated antigen (mutated) 1-like 1 [Source:HGNC Symbol;Acc:26583] REFSEQ_MRNA 12 NM_002923.3 12.1 ENSG00000116741 RGS2 regulator of G-protein signaling 2, 24kDa [Source:HGNC Symbol;Acc:9998] REFSEQ_MRNA 13 NR_003038.2 13.1 N/A N/A N/A N/A 14 NM_080862.1 14.1 ENSG00000175093 SPSB4 splA/ryanodine receptor -

Gene List of the Targeted NGS MCD and CCA Gene Panel AKT3,ALX1
Gene List of the targeted NGS MCD and CCA gene panel AKT3,ALX1,ALX3,ALX4,AMPD2,ARFGEF2,ARID1B,ARX,ASPM,ATR,ATRX,B3GALTL,BRPF1,c12orf57,C6orf70,CASK,CCND2,CDK5RAP2,CDON,C ENPJ,CEP170,CHMP1A,COL4A1,CREBBP,CYP11A1,DCHS1,DCLK1,DCX,DHCR24,DHCR7,DIS3L2,DISC1,DISP1,DLL1,DMRTA2,DYNC1H1,DYRK1 A,EARS2,EFNB1,EMX1,EOMES,EP300,ERBB4,ERMARD,EXOSC3,FAM36A,FGF8,FGFR1,FGFR2,FLNA,FOXC1,FOXG1,FOXH1,FZD10,GLI2,GLI3,GP R56,GPSM2,HCCS,HESX1,HNRNPU,IGBP1,IGFBP1,ISPD,ITPA,KAL1,KAT6B,KATNB1,KIAA1279,KIF14,KIF1A,KIF1B,KIF21A,KIF2A,KIF5C,KIF7,L1 CAM,LAMB1,LAMC3,LRP2,MCPH1,MED12,MID1,NDE1,NFIB,NPC1,NR2F1,NSD1,NTRK1,NTRK3,OCEL1,OPA1,OTX2,PAFAH1B1,PAX6,PEX1,PHF1 0,PIK3R2,POLR3A,POLR3B,POMT1,POMT2,PTCH1,PTPRS,PYCR1,RAB3GAP1,RARS2,RELN,RFX3,ROBO1,ROBO3,RPS6KA3,RTTN,SATB2,SEPSEC S,SHH,SIX3,SLC12A6,SOX2,SPOCK1,SRPX2,TBCD,TBCE,TCF4,TDGF1,TEAD1,THBS2,TMEM5,TSC1,TSC2,TSEN15,TSEN2,TSEN34,TSEN54,TUBA1 A,TUBA8,TUBB,TUBB2A,TUBB2B,TUBB3,TUBB4A,TUBG1,VAX1,VRK1,WDR47,WDR62,ZBTB18,ZEB2,ZIC2. Gene List of the targeted NGS epilepsy gene panel AARS, ADGRV1, ADRA2B, ADSL, ALDH4A1, ALDH7A1, ALG13, ALPL, ARHGEF15, ARHGEF9, ARX, ASAH1, ATP1A2, ATP1A3, BRD2, CACNA1A, CACNA1H, CACNA2D2, CACNB4, CBL, CDKL5, CERS1, CHD2, CHRNA2, CHRNA4, CHRNB2, CLCN2, CLCN4, CLN8, CLTC, CNKSR2, CNTNAP2, CPA6, CPLX1, CSNK1G1, CSNK2B, CTNND2, DEPDC5, DHDDS, DNM1, DOCK7, DYNC1H1, EEF1A2, EFHC1, EIF2S3, EMC1, EPM2A, FASN, FLNA, FOXG1, GABBR2, GABRA1, GABRA2, GABRA3, GABRB2, GABRB3, GABRD, GABRG2, GAL, GNAO1, GOSR2, GRIA1, GRIN1, GRIN2A, GRIN2B, HCN1, HCN4, HDAC4, HNRNPU, IDH3A, IQSEC2, JRK, KCNA1, KCNA2, KCNB1, -

CENTOGENE's Severe and Early Onset Disorder Gene List
CENTOGENE’s severe and early onset disorder gene list USED IN PRENATAL WES ANALYSIS AND IDENTIFICATION OF “PATHOGENIC” AND “LIKELY PATHOGENIC” CENTOMD® VARIANTS IN NGS PRODUCTS The following gene list shows all genes assessed in prenatal WES tests or analysed for P/LP CentoMD® variants in NGS products after April 1st, 2020. For searching a single gene coverage, just use the search on www.centoportal.com AAAS, AARS1, AARS2, ABAT, ABCA12, ABCA3, ABCB11, ABCB4, ABCB7, ABCC6, ABCC8, ABCC9, ABCD1, ABCD4, ABHD12, ABHD5, ACACA, ACAD9, ACADM, ACADS, ACADVL, ACAN, ACAT1, ACE, ACO2, ACOX1, ACP5, ACSL4, ACTA1, ACTA2, ACTB, ACTG1, ACTL6B, ACTN2, ACVR2B, ACVRL1, ACY1, ADA, ADAM17, ADAMTS2, ADAMTSL2, ADAR, ADARB1, ADAT3, ADCY5, ADGRG1, ADGRG6, ADGRV1, ADK, ADNP, ADPRHL2, ADSL, AFF2, AFG3L2, AGA, AGK, AGL, AGPAT2, AGPS, AGRN, AGT, AGTPBP1, AGTR1, AGXT, AHCY, AHDC1, AHI1, AIFM1, AIMP1, AIPL1, AIRE, AK2, AKR1D1, AKT1, AKT2, AKT3, ALAD, ALDH18A1, ALDH1A3, ALDH3A2, ALDH4A1, ALDH5A1, ALDH6A1, ALDH7A1, ALDOA, ALDOB, ALG1, ALG11, ALG12, ALG13, ALG14, ALG2, ALG3, ALG6, ALG8, ALG9, ALMS1, ALOX12B, ALPL, ALS2, ALX3, ALX4, AMACR, AMER1, AMN, AMPD1, AMPD2, AMT, ANK2, ANK3, ANKH, ANKRD11, ANKS6, ANO10, ANO5, ANOS1, ANTXR1, ANTXR2, AP1B1, AP1S1, AP1S2, AP3B1, AP3B2, AP4B1, AP4E1, AP4M1, AP4S1, APC2, APTX, AR, ARCN1, ARFGEF2, ARG1, ARHGAP31, ARHGDIA, ARHGEF9, ARID1A, ARID1B, ARID2, ARL13B, ARL3, ARL6, ARL6IP1, ARMC4, ARMC9, ARSA, ARSB, ARSL, ARV1, ARX, ASAH1, ASCC1, ASH1L, ASL, ASNS, ASPA, ASPH, ASPM, ASS1, ASXL1, ASXL2, ASXL3, ATAD3A, ATCAY, ATIC, ATL1, ATM, ATOH7, -

Gene Trapping Identifies Chloride Channel 4 As a Novel Inducer of Colon Cancer Cell Migration, Invasion and Metastases
British Journal of Cancer (2010) 102, 774 – 782 & 2010 Cancer Research UK All rights reserved 0007 – 0920/10 $32.00 www.bjcancer.com Gene trapping identifies chloride channel 4 as a novel inducer of colon cancer cell migration, invasion and metastases 1 1 2 3 4 *,1 T Ishiguro , H Avila , S-Y Lin , T Nakamura , M Yamamoto and DD Boyd 1 2 Cancer Biology Department, MD Anderson Cancer Center, Houston, TX, USA; Systems Biology Department, MD Anderson Cancer Center, Houston, 3 4 TX, USA; Surgical Oncology Department, Hokkaido University Graduate School of Medicine, Sapporo, Japan; Second Department of Surgery, Hamamatsu University School of Medicine, Hamanatsu, Japan BACKGROUND: To date, there are few reports on gene products contributing to colon cancer progression. METHODS: We used a gene trap comprised of an enhanced retroviral mutagen (ERM) cassette that includes a tetracycline-responsive promoter upstream of a haemagglutinin (HA) tag and a splice donor site. Integration of the ERM within an endogenous gene yields a tetracycline-regulated HA-tagged transcript. We transduced RKO colon cancer cells expressing a tetracycline trans-activator-off with the ERM-encoding retrovirus and screened for enhanced migration. RESULTS: One clone showed fivefold enhanced migration with tetracycline withdrawal. Rapid amplification of cDNA ends identified the trapped gene as the chloride channel 4 (CLCN4) exchanger. Stable expression of a CLCN4 cDNA enhanced motility, whereas cells knocked down or null for this transcript showed reduced migration/invasion. CLCN4-overexpressing RKO colon cancer cells were þ more resistant than controls to proton load-induced cytotoxicity, consistent with the H -extruding function of this antiporter. -

NIH Public Access Author Manuscript FEBS Lett
NIH Public Access Author Manuscript FEBS Lett. Author manuscript; available in PMC 2011 May 17. NIH-PA Author ManuscriptPublished NIH-PA Author Manuscript in final edited NIH-PA Author Manuscript form as: FEBS Lett. 2010 May 17; 584(10): 2102±2111. doi:10.1016/j.febslet.2010.01.037. Chloride Channels of Intracellular Membranes John C. Edwards* and Christina R. Kahl UNC Kidney Center and the Division of Nephrology and Hypertension, Department of Medicine, University of North Carolina at Chapel Hill Abstract Proteins implicated as intracellular chloride channels include the intracellular ClC proteins, the bestrophins, the cystic fibrosis transmembrane conductance regulator, the CLICs, and the recently described Golgi pH regulator. This paper examines current hypotheses regarding roles of intracellular chloride channels and reviews the evidence supporting a role in intracellular chloride transport for each of these proteins. Keywords chloride channel; ClC; CLIC; bestrophin; GPHR The study of chloride channels of intracellular membranes has seen enormous advances over the past two decades and exciting recent developments have sparked renewed interest in this field. The discovery of important roles for intracellular chloride channels in human disease processes as diverse as retinal macular dystrophy, osteopetrosis, renal proximal tubule dysfunction, and angiogenesis have highlighted the importance of these molecules in critical cellular activities. Startling discoveries regarding the intracellular ClC family of proteins have forced a re-examination -

1 1 2 3 Cell Type-Specific Transcriptomics of Hypothalamic
1 2 3 4 Cell type-specific transcriptomics of hypothalamic energy-sensing neuron responses to 5 weight-loss 6 7 Fredrick E. Henry1,†, Ken Sugino1,†, Adam Tozer2, Tiago Branco2, Scott M. Sternson1,* 8 9 1Janelia Research Campus, Howard Hughes Medical Institute, 19700 Helix Drive, Ashburn, VA 10 20147, USA. 11 2Division of Neurobiology, Medical Research Council Laboratory of Molecular Biology, 12 Cambridge CB2 0QH, UK 13 14 †Co-first author 15 *Correspondence to: [email protected] 16 Phone: 571-209-4103 17 18 Authors have no competing interests 19 1 20 Abstract 21 Molecular and cellular processes in neurons are critical for sensing and responding to energy 22 deficit states, such as during weight-loss. AGRP neurons are a key hypothalamic population 23 that is activated during energy deficit and increases appetite and weight-gain. Cell type-specific 24 transcriptomics can be used to identify pathways that counteract weight-loss, and here we 25 report high-quality gene expression profiles of AGRP neurons from well-fed and food-deprived 26 young adult mice. For comparison, we also analyzed POMC neurons, an intermingled 27 population that suppresses appetite and body weight. We find that AGRP neurons are 28 considerably more sensitive to energy deficit than POMC neurons. Furthermore, we identify cell 29 type-specific pathways involving endoplasmic reticulum-stress, circadian signaling, ion 30 channels, neuropeptides, and receptors. Combined with methods to validate and manipulate 31 these pathways, this resource greatly expands molecular insight into neuronal regulation of 32 body weight, and may be useful for devising therapeutic strategies for obesity and eating 33 disorders. -

Application of Microarrays to Neurological Disease
BASIC SCIENCE SEMINARS IN NEUROLOGY SECTION EDITOR: HASSAN M. FATHALLAH-SHAYKH, MD Application of Microarrays to Neurological Disease Lisa-Marie Sturla, PhD; Ana Fernandez-Teijeiro, MD, PhD; Scott L. Pomeroy, MD, PhD odern microarray-based functional genomics holds great promise for revealing novel molecular and cellular mechanisms of disease. First introduced commercially in 1996, microarrays have been used widely to monitor the expression of thousands of genes in biological samples, as described in the following paragraphs. Other mi- Mcroarray-based genomic applications are also in development, including comparative genomic hy- bridization, on-chip sequencing, and novel drug discovery. For example, DNA array-based com- parative genomic hybridization identifies chromosomal gains and losses with greatly improved resolution compared with conventional methods that use metaphase chromosomes as hybridiza- tion targets.1 This increase in resolution will continue to improve as the technology advances. More- over, microarrays provide a better platform for automation than is possible with standard meta- phase techniques. Where genetic mutations and aberrations are already well characterized, microarrays can be customized to be effectively used as a diagnostic and prognostic tool.2,3 In the field of drug discovery, microarrays have the potential to dramatically enhance progress, being used at all stages from target discovery (through validation of new molecular targets and understanding modes of action) to predicting patient response.4 These devices are beginning to revolu- the application of microarray technology tionize how scientists explore the opera- and emerging data analysis techniques to tion of normal cells in the body and the pediatric brain tumors.8 Using microar- molecular aberrations that underlie medi- rays that monitor the expression of more cal disorders. -

Transdifferentiation of Human Mesenchymal Stem Cells
Transdifferentiation of Human Mesenchymal Stem Cells Dissertation zur Erlangung des naturwissenschaftlichen Doktorgrades der Julius-Maximilians-Universität Würzburg vorgelegt von Tatjana Schilling aus San Miguel de Tucuman, Argentinien Würzburg, 2007 Eingereicht am: Mitglieder der Promotionskommission: Vorsitzender: Prof. Dr. Martin J. Müller Gutachter: PD Dr. Norbert Schütze Gutachter: Prof. Dr. Georg Krohne Tag des Promotionskolloquiums: Doktorurkunde ausgehändigt am: Hiermit erkläre ich ehrenwörtlich, dass ich die vorliegende Dissertation selbstständig angefertigt und keine anderen als die von mir angegebenen Hilfsmittel und Quellen verwendet habe. Des Weiteren erkläre ich, dass diese Arbeit weder in gleicher noch in ähnlicher Form in einem Prüfungsverfahren vorgelegen hat und ich noch keinen Promotionsversuch unternommen habe. Gerbrunn, 4. Mai 2007 Tatjana Schilling Table of contents i Table of contents 1 Summary ........................................................................................................................ 1 1.1 Summary.................................................................................................................... 1 1.2 Zusammenfassung..................................................................................................... 2 2 Introduction.................................................................................................................... 4 2.1 Osteoporosis and the fatty degeneration of the bone marrow..................................... 4 2.2 Adipose and bone -

CLCN4 Polyclonal Antibody
CLCN4 polyclonal antibody Catalog # : PAB22540 規格 : [ 100 uL ] List All Specification Application Image Product Rabbit polyclonal antibody raised against recombinant CLCN4. Immunohistochemistry (Formalin/PFA-fixed paraffin- Description: embedded sections) Immunogen: Recombinant protein corresponding to amino acids of human CLCN4. Sequence: VVSRDSERLIGFAQRRELILAIKNARQRQEGIVSNSIMYFTEEPPELPANSP HPLKLRRILNLS Host: Rabbit enlarge Reactivity: Human Form: Liquid Purification: Antigen affinity purification Isotype: IgG Recommend Immunohistochemistry (1:20-1:50) Usage: The optimal working dilution should be determined by the end user. Storage Buffer: In PBS, pH 7.2 (40% glycerol, 0.02% sodium azide) Storage Store at 4°C. For long term storage store at -20°C. Instruction: Aliquot to avoid repeated freezing and thawing. Note: This product contains sodium azide: a POISONOUS AND HAZARDOUS SUBSTANCE which should be handled by trained staff only. Datasheet: Download Applications Immunohistochemistry (Formalin/PFA-fixed paraffin-embedded sections) Immunohistochemical staining of human pancreas with CLCN4 polyclonal antibody (Cat # PAB22540) shows strong cytoplasmic positivity in exocrine cells. Gene Information Page 1 of 2 2019/4/9 Entrez GeneID: 1183 Protein P51793 Accession#: Gene Name: CLCN4 Gene Alias: CLC4,ClC-4,ClC-4A,MGC163150 Gene chloride channel 4 Description: Omim ID: 302910 Gene Ontology: Hyperlink Gene Summary: The CLCN family of voltage-dependent chloride channel genes comprises nine members (CLCN1-7, Ka and Kb) which demonstrate quite diverse functional characteristics while sharing significant sequence homology. Chloride channel 4 has an evolutionary conserved CpG island and is conserved in both mouse and hamster. This gene is mapped in close proximity to APXL (Apical protein Xenopus laevis-like) and OA1 (Ocular albinism type I), which are both located on the human X chromosome at band p22.3.