10.8 Transduction in Transduction, a Bacterial Virus (Bacteriophage) Transfers DNA from One Cell to Another
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Topic: Genetic Recombination of Bacteria
COMPILED AND CIRCULATED BY ARPITA CHAKRABORTY, STATE AIDED COLLEGE TEACHER, DEPARTMENT OF BOTANY, NARAJOLE RAJ COLLEGE. Topic: Genetic Recombination of Bacteria Genetic Recombination of Bacteria: The following points highlight the three main processes involved in the genetic recombination of bacteria. The processes are: 1. Conjugation 2. Transformation 3. Transduction. Process- 1. Conjugation: In this process, the exchange of genetic material takes place through a conjugation tube between the two cells of bacteria. The process was first postulated by Joshua Lederberg and Edward Tatum (1946) in Escherichia coli. They were awarded the Nobel Prize in 1958 for their work on bacterial genetics. Later on, it has also been demonstrated in Salmonella, Vibrio and Pseudomonas. There are two mating types of bacteria, one is. male type or F+ or donor cell, which donates some DNA. The other one is female type or F– or recipient cell, which receives DNA. Later, after receiving DNA, the recipient cell may behave as donor cell i.e., F+ type. The F-factor is the fertility factor, sex-factor or F-plasmid present in the cell of F+ i.e., donor cell or male type. The plasmid takes part in conjugation is called episome. BOTANY: SEM- I, PAPER: C1T:PHYCOLOGY AND MICROBIOLOGY, UNIT 3: BACTERIA COMPILED AND CIRCULATED BY ARPITA CHAKRABORTY, STATE AIDED COLLEGE TEACHER, DEPARTMENT OF BOTANY, NARAJOLE RAJ COLLEGE. In this process, two cells of opposite mating type i.e., F+ and F– become temporarily attached with each other by sex pilus. The sex pilus has a hole of 2.5 pm diameter through which DNA can pass from donor to recipient cell. -

Mobile Genetic Elements in Streptococci
Curr. Issues Mol. Biol. (2019) 32: 123-166. DOI: https://dx.doi.org/10.21775/cimb.032.123 Mobile Genetic Elements in Streptococci Miao Lu#, Tao Gong#, Anqi Zhang, Boyu Tang, Jiamin Chen, Zhong Zhang, Yuqing Li*, Xuedong Zhou* State Key Laboratory of Oral Diseases, National Clinical Research Center for Oral Diseases, West China Hospital of Stomatology, Sichuan University, Chengdu, PR China. #Miao Lu and Tao Gong contributed equally to this work. *Address correspondence to: [email protected], [email protected] Abstract Streptococci are a group of Gram-positive bacteria belonging to the family Streptococcaceae, which are responsible of multiple diseases. Some of these species can cause invasive infection that may result in life-threatening illness. Moreover, antibiotic-resistant bacteria are considerably increasing, thus imposing a global consideration. One of the main causes of this resistance is the horizontal gene transfer (HGT), associated to gene transfer agents including transposons, integrons, plasmids and bacteriophages. These agents, which are called mobile genetic elements (MGEs), encode proteins able to mediate DNA movements. This review briefly describes MGEs in streptococci, focusing on their structure and properties related to HGT and antibiotic resistance. caister.com/cimb 123 Curr. Issues Mol. Biol. (2019) Vol. 32 Mobile Genetic Elements Lu et al Introduction Streptococci are a group of Gram-positive bacteria widely distributed across human and animals. Unlike the Staphylococcus species, streptococci are catalase negative and are subclassified into the three subspecies alpha, beta and gamma according to the partial, complete or absent hemolysis induced, respectively. The beta hemolytic streptococci species are further classified by the cell wall carbohydrate composition (Lancefield, 1933) and according to human diseases in Lancefield groups A, B, C and G. -

Horizontal Gene Transfer
Genetic Variation: The genetic substrate for natural selection Horizontal Gene Transfer Dr. Carol E. Lee, University of Wisconsin Copyright ©2020; Do not upload without permission What about organisms that do not have sexual reproduction? In prokaryotes: Horizontal gene transfer (HGT): Also termed Lateral Gene Transfer - the lateral transmission of genes between individual cells, either directly or indirectly. Could include transformation, transduction, and conjugation. This transfer of genes between organisms occurs in a manner distinct from the vertical transmission of genes from parent to offspring via sexual reproduction. These mechanisms not only generate new gene assortments, they also help move genes throughout populations and from species to species. HGT has been shown to be an important factor in the evolution of many organisms. From some basic background on prokaryotic genome architecture Smaller Population Size • Differences in genome architecture (noncoding, nonfunctional) (regulatory sequence) (transcribed sequence) General Principles • Most conserved feature of Prokaryotes is the operon • Gene Order: Prokaryotic gene order is not conserved (aside from order within the operon), whereas in Eukaryotes gene order tends to be conserved across taxa • Intron-exon genomic organization: The distinctive feature of eukaryotic genomes that sharply separates them from prokaryotic genomes is the presence of spliceosomal introns that interrupt protein-coding genes Small vs. Large Genomes 1. Compact, relatively small genomes of viruses, archaea, bacteria (typically, <10Mb), and many unicellular eukaryotes (typically, <20 Mb). In these genomes, protein-coding and RNA-coding sequences occupy most of the genomic sequence. 2. Expansive, large genomes of multicellular and some unicellular eukaryotes (typically, >100 Mb). In these genomes, the majority of the nucleotide sequence is non-coding. -

Transduction by Bacteriophage Ti * Henry Drexler
Proceedings of the National Academy of Sciences Vol. 66, No. 4, pp. 1083-1088, August 1970 Transduction by Bacteriophage Ti * Henry Drexler DEPARTMENT OF MICROBIOLOGY, BOWMAN GRAY SCHOOL OF MEDICINE, WAKE FOREST UNIVERSITY, WINSTON-SALEM, NORTH CAROLINA Communicated by Edward L. Tatum, May 25, 1970 Abstract. Amber mutants of the virulent coliphage T1 are able to transduce a wide variety of genetic characteristics from permissive to nonpermissive K strains of Escherichia coli. The virulent coliphage T1 is not known to be related to any temperate phage. Certain of the characteristics of T1 seem to be incompatible with its potential existence as a temperate phage; for example, the average latent period for T1 is only 13 min' and about 70% of its DNA is derived from the host.2 In order to demonstrate transduction by T1 it is necessray to provide condi- tions in which potential transductants are able to survive; this is accomplished by using amber mutants of T1 to transduce nonpermissive recipients. Experi- ments which show that T1 is a transducing phage are presented and have been designed chiefly to illustrate the following points: (1) Infection by T1 is able to cause heritable changes in recipients; (2) the genotype of the donor host is im- portant in determining what characteristics can be transferred to recipients; (3) the changes in the recipients are not caused by transformation; and (4) there is a similarity between the transducing activity and the plaque-forming ability of T1 with respect to serology, host range, and density. Materials and Methods. Bacterial strains: The abbreviations and symbols of Demerec et al.3 and Taylor and Trotter4 are used to describe all pertinent genotypes. -

Bacterial Genetics
BACTERIAL GENETICS Genetics is the study of genes including the structure of genetic materials, what information is stored in the genes, how the genes are expressed and how the genetic information is transferred. Genetics is also the study of heredity and variation. The arrangement of genes within organisms is its genotype and the physical characteristics an organism based on its genotype and the interaction with its environment, make up its phenotype. The order of DNA bases constitutes the bacterium's genotype. A particular organism may possess alternate forms of some genes. Such alternate forms of genes are referred to as alleles. The cell's genome is stored in chromosomes, which are chains of double stranded DNA. Genes are sequences of nucleotides within DNA that code for functional proteins. The genetic material of bacteria and plasmids is DNA. The two essential functions of genetic material are replication and expression. Structure of DNA The DNA molecule is composed of two chains of nucleotides wound around each other in the form of “double helix”. Double-stranded DNA is helical, and the two strands in the helix are antiparallel. The backbone of each strand comprises of repeating units of deoxyribose and phosphate residue. Attached to the deoxyribose is purine (AG) or pyrimidine (CT) base. Nucleic acids are large polymers consisting of repeating nucleotide units. Each nucleotide contains one phosphate group, one deoxyribose sugar, and one purine or pyrimidine base. In DNA the sugar is deoxyribose; in RNA the sugar is ribose. The double helix is stabilized by hydrogen bonds between purine and pyrimidine bases on the opposite strands. -

Recombination in Bacteria
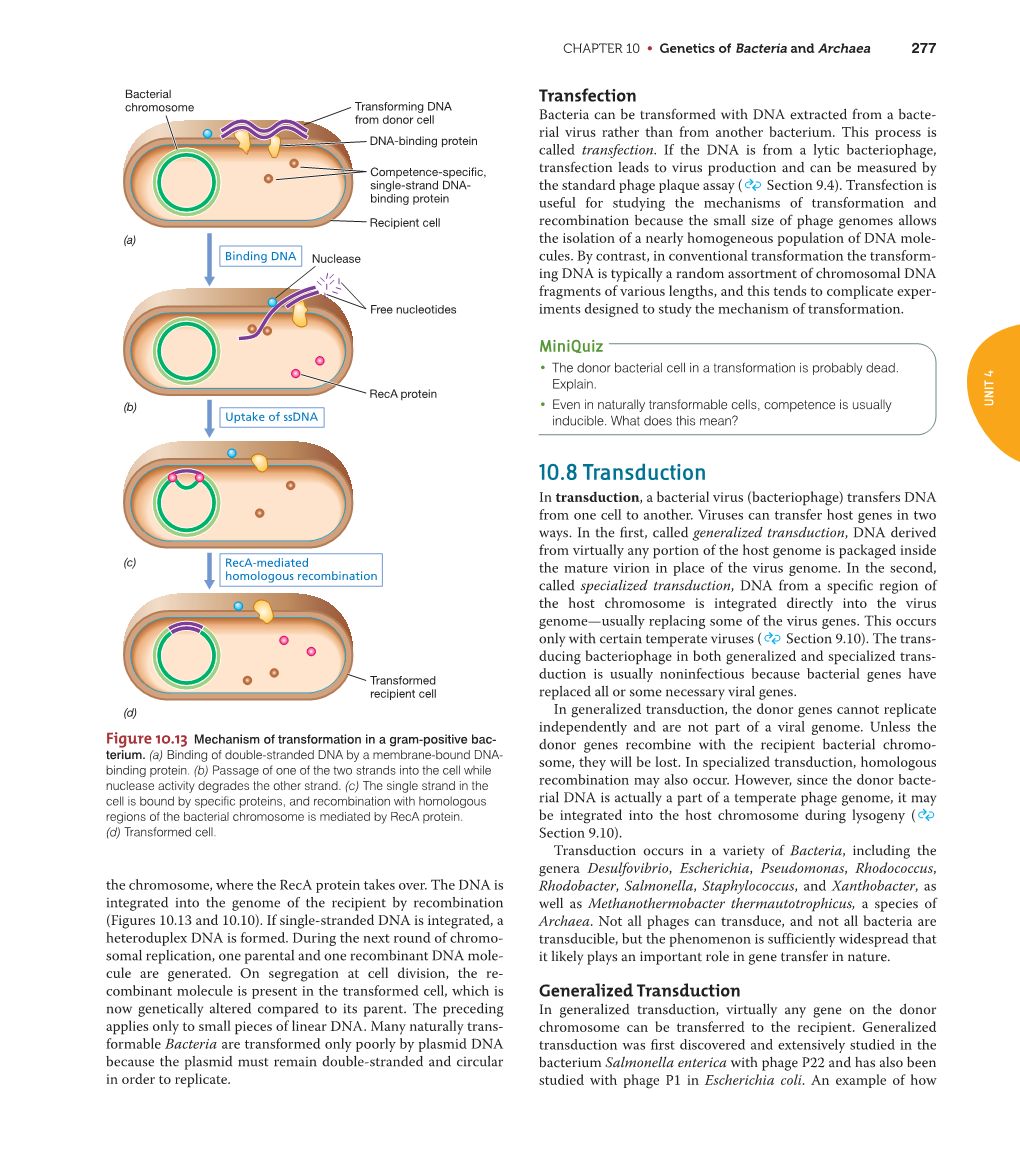
Recombination in Bacteria 1. Conjugation DNA from a donor cell is transferred to a recipient cell through a conjugation tube (pili). 2. Transformation Uptake of naked DNA molecule from remains of one bacterium (donor cell) by another bacterium (recipient cell). 3. Transduction Bacterial genes are carried from a donor cell to a recipient cell by a bacteriophage. { Generalized { Specialized Conjugation Ability to conjugate located on the F-plasmid F+ Cells act as donors F- Cells act as recipients F+/F- Conjugation: { F Factor “replicates off” a single strand of DNA. { New strand goes through pili to recipient cell. { New strand is made double stranded. { If entire F-plasmid crosses, then recipient cell becomes F+, otherwise nothing happens Conjugation with Hfr Hfr cell (High Frequency Recombination) cells have F-plasmid integrated into the Chromosome. Integration into the Chromosome is unique for each F-plasmid strain. When F-plasmid material is replicated and sent across pili, Chromosomal material is included. (Figure 6.10 in Klug & Cummings) When chromosomal material is in recipient cell, recombination can occur: { Recombination is double stranded. { Donor genes are recombined into the recipient cell. { Corresponding genes from recipient cell are recombined out of the chromosome and reabsorbed by the cell. Interrupted Mating Mapping 1. Allow conjugation to start Genes closest to the origin of replication site (in the direction of replication) are moved through the pili first. 2. After a set time, interrupt conjugation Only those genes closest to the origin of replication site will conjugate. The long the time, the more that is able to conjugate. 3. -

Microbial Genetics by Dr Preeti Bajpai
Dr. Preeti Bajpai Genes: an overview ▪ A gene is the functional unit of heredity ▪ Each chromosome carry a linear array of multiple genes ▪ Each gene represents segment of DNA responsible for synthesis of RNA or protein product ▪ A gene is considered to be unit of genetic information that controls specific aspect of phenotype DNA Chromosome Gene Protein-1 Prokaryotic Courtesy: Team Shrub https://twitter.com/realscientists/status/927 cell 667237145767937 Genetic exchange within Prokaryotes The genetic exchange occurring in bacteria involve transfers of genes from one bacterium to another. The gene transfer in prokaryotic cells is thus unidirectional and the recombination events usually occur between a fragment of one chromosome (from a donor cell) and a complete chromosome (in a recipient cell) Mechanisms for genetic exchange Bacteria exchange genetic material through three different parasexual processes* namely transformation, conjugation and transduction. *Parasexual process involves recombination of genes from genetically distinct cells occurring without involvement of meiosis and fertilization Principles of Genetics-sixth edition Courtesy: Beatrice the Biologist.com by D. Peter Snustad & Michael J. Simmons (http://www.beatricebiologist.com/2014/08/bacterial-gifts/) Transformation: an introduction Transformation involves the uptake of free DNA molecules released from one bacterium (the donor cell) by another bacterium (the recipient cell). Frederick Griffith discovered transformation in Streptococcus pneumoniae (pneumococcus) in 1928. In his experiments, Griffith used two related strains of bacteria, known as R and S. The R bacteria (nonvirulent) formed colonies, or clumps of related bacteria, that Frederick Griffith 1877-1941 had a rough appearance (hence the abbreviation "R"). The S bacteria (virulent) formed colonies that were rounded and smooth (hence the abbreviation "S"). -

Comprehensive Analysis of Mobile Genetic Elements in the Gut Microbiome Reveals Phylum-Level Niche-Adaptive Gene Pools
bioRxiv preprint doi: https://doi.org/10.1101/214213; this version posted December 22, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Comprehensive analysis of mobile genetic elements in the gut microbiome 2 reveals phylum-level niche-adaptive gene pools 3 Xiaofang Jiang1,2,†, Andrew Brantley Hall2,3,†, Ramnik J. Xavier1,2,3,4, and Eric Alm1,2,5,* 4 1 Center for Microbiome Informatics and Therapeutics, Massachusetts Institute of Technology, 5 Cambridge, MA 02139, USA 6 2 Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA 7 3 Center for Computational and Integrative Biology, Massachusetts General Hospital and Harvard 8 Medical School, Boston, MA 02114, USA 9 4 Gastrointestinal Unit and Center for the Study of Inflammatory Bowel Disease, Massachusetts General 10 Hospital and Harvard Medical School, Boston, MA 02114, USA 11 5 MIT Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA 12 02142, USA 13 † Co-first Authors 14 * Corresponding Author bioRxiv preprint doi: https://doi.org/10.1101/214213; this version posted December 22, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 15 Abstract 16 Mobile genetic elements (MGEs) drive extensive horizontal transfer in the gut microbiome. This transfer 17 could benefit human health by conferring new metabolic capabilities to commensal microbes, or it could 18 threaten human health by spreading antibiotic resistance genes to pathogens. Despite their biological 19 importance and medical relevance, MGEs from the gut microbiome have not been systematically 20 characterized. -

Molecular Model for the Transposition and Replication of Bacteriophage
Proc. Natl. Acad. Sci. USA Vol. 76, No. 4, pp. 1933-1937, April 1979 Genetics Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements (DNA insertion elements/nonhomologous recombination/site-specific recombination/replicon fusion/topoisomerases) JAMES A. SHAPIRO Department of Microbiology, The University of Chicago, Chicago, Illinois 60637 Communicated by Hewson Swift, December 11, 1978 ABSTRACT A series of molecular events will explain how B genetic elements can transpose from one DNA site to another, B y generate a short oligonucleotide duplication at both ends of the I % new insertion site, and replicate in the transposition process. I These events include the formation of recombinant molecules A a% /; C which have been postulated to be intermediates in the trans- position process. The model explains how the replication of bacteriophage Mu is obligatorily associated with movement to z x x new genetic sites. It postulates that all transposable elements replicate in the transposition process so that they remain at their z original site while moving to new sites. According to this model, the mechanism of transposition is very different from the in- V sertion and excision of bacteriophage X. y FIG. 1. Replicon fusion. The boxes with an arrow indicate copies Recent research on transposable elements in bacteria has pro- of a transposable element. The letters mark arbitrary regions of the vided important insights into the role of nonhomologous re- two replicons to indicate the relative positions of the elements in the combination in genetic rearrangements (1-4). These elements donor and cointegrate molecules. include small insertion sequences (IS elements), transposable resistance determinants (Tn elements), and bacteriophage Mu results in the duplication of a short oligonucleotide sequence (3). -

Section 4. Guidance Document on Horizontal Gene Transfer Between Bacteria
306 - PART 2. DOCUMENTS ON MICRO-ORGANISMS Section 4. Guidance document on horizontal gene transfer between bacteria 1. Introduction Horizontal gene transfer (HGT) 1 refers to the stable transfer of genetic material from one organism to another without reproduction. The significance of horizontal gene transfer was first recognised when evidence was found for ‘infectious heredity’ of multiple antibiotic resistance to pathogens (Watanabe, 1963). The assumed importance of HGT has changed several times (Doolittle et al., 2003) but there is general agreement now that HGT is a major, if not the dominant, force in bacterial evolution. Massive gene exchanges in completely sequenced genomes were discovered by deviant composition, anomalous phylogenetic distribution, great similarity of genes from distantly related species, and incongruent phylogenetic trees (Ochman et al., 2000; Koonin et al., 2001; Jain et al., 2002; Doolittle et al., 2003; Kurland et al., 2003; Philippe and Douady, 2003). There is also much evidence now for HGT by mobile genetic elements (MGEs) being an ongoing process that plays a primary role in the ecological adaptation of prokaryotes. Well documented is the example of the dissemination of antibiotic resistance genes by HGT that allowed bacterial populations to rapidly adapt to a strong selective pressure by agronomically and medically used antibiotics (Tschäpe, 1994; Witte, 1998; Mazel and Davies, 1999). MGEs shape bacterial genomes, promote intra-species variability and distribute genes between distantly related bacterial genera. Horizontal gene transfer (HGT) between bacteria is driven by three major processes: transformation (the uptake of free DNA), transduction (gene transfer mediated by bacteriophages) and conjugation (gene transfer by means of plasmids or conjugative and integrated elements). -

The Obscure World of Integrative and Mobilizable Elements Gérard Guédon, Virginie Libante, Charles Coluzzi, Sophie Payot-Lacroix, Nathalie Leblond-Bourget
The obscure world of integrative and mobilizable elements Gérard Guédon, Virginie Libante, Charles Coluzzi, Sophie Payot-Lacroix, Nathalie Leblond-Bourget To cite this version: Gérard Guédon, Virginie Libante, Charles Coluzzi, Sophie Payot-Lacroix, Nathalie Leblond-Bourget. The obscure world of integrative and mobilizable elements: Highly widespread elements that pirate bacterial conjugative systems. Genes, MDPI, 2017, 8 (11), pp.337. 10.3390/genes8110337. hal- 01686871 HAL Id: hal-01686871 https://hal.archives-ouvertes.fr/hal-01686871 Submitted on 26 May 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License G C A T T A C G G C A T genes Review The Obscure World of Integrative and Mobilizable Elements, Highly Widespread Elements that Pirate Bacterial Conjugative Systems Gérard Guédon *, Virginie Libante, Charles Coluzzi, Sophie Payot and Nathalie Leblond-Bourget * ID DynAMic, Université de Lorraine, INRA, 54506 Vandœuvre-lès-Nancy, France; [email protected] (V.L.); [email protected] (C.C.); [email protected] (S.P.) * Correspondence: [email protected] (G.G.); [email protected] (N.L.-B.); Tel.: +33-037-274-5142 (G.G.); +33-037-274-5146 (N.L.-B.) Received: 12 October 2017; Accepted: 15 November 2017; Published: 22 November 2017 Abstract: Conjugation is a key mechanism of bacterial evolution that involves mobile genetic elements. -

Genetic Exchange in Bacteria
Systems Microbiology Monday Oct 16 - Ch 10 -Brock Genetic Exchange in Bacteria •• HomologousHomologous recombinationrecombination •• TransformationTransformation •• PlasmidsPlasmids andand conjugationconjugation •• TransposableTransposable elementselements •• TransductionTransduction (virus(virus mediatedmediated xchangexchange)) Gene exchange in bacteria • Transfer of DNA from one bacterium to another is a common means of gene dispersal. It has a big effect on bacterial evolution, and tremendous practical implications. For example, lateral transfer is responsible for the spread drug resistance determinants between bacterial species. • Three common mechanisms of lateral gene exchange : – Transformation (extracellular DNA uptake) – Conjugation (bacterial mating systems) – Transduction (viral mediated gene exchange) RecA mediated Homologous recombination Images removed due to copyright restrictions. See Figures 10-9 and 10-10 in Madigan, Michael, and John Martinko. Brock Biology of Microorganisms.11th ed. Upper Saddle River, NJ: Pearson Prentice Hall, 2006. ISBN: 0131443291. Gene exchange in bacteriaTransformation The Griffith Experiment S Injection Dead mouse; • Discovered by Griffith in yields S1 cells 1928 during the course of his Live "smooth" (encapsulated) studies of virulence in type 1 pneumococci (S1) Streptococcus pneumoniae. Dead S Live mouse Heat-killed S1 • S=smooth colony Live mouse morphotype Live "rough" (unencapsulated) pneumococci (R1 or R2) derived by subculture from S1 or S2, respectively • R=rough colony Dead mouse; yields S cells morphotype R1 + Dead S + 1 Live R1 Killed S1 R2 + Dead S + Dead mouse; yields S1 cells Live R2 Killed S2 Figure by MIT OCW. Gene exchange mechanisms in bacteria Transformation The Griffith Experiment Injection Dead mouse; S yields S cells Avery, MacLeod, and McCarthy (1944) 1 fractionation studies led to conclusion that Live "smooth" (encapsulated) type 1 pneumococci (S ) transformation principle is DNA.