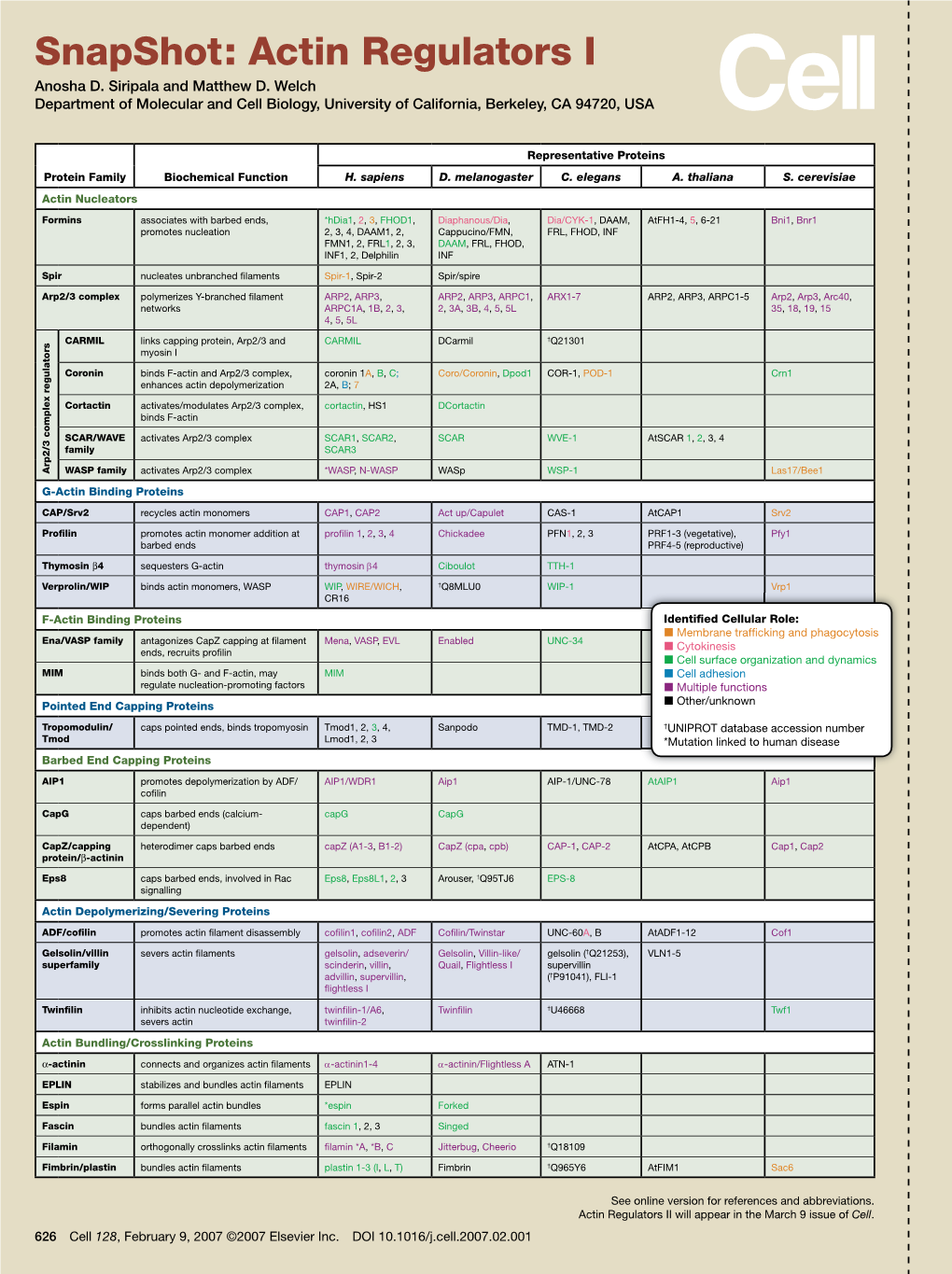
Snapshot: Actin Regulators I Anosha D
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Large-Scale Opening of Utrophints Tandem Calponin Homology (CH
Large-scale opening of utrophin’s tandem calponin homology (CH) domains upon actin binding by an induced-fit mechanism Ava Y. Lin, Ewa Prochniewicz, Zachary M. James, Bengt Svensson, and David D. Thomas1 Department of Biochemistry, Molecular Biology and Biophysics, University of Minnesota, Minneapolis, MN 55455 Edited by James A. Spudich, Stanford University School of Medicine, Stanford, CA, and approved June 20, 2011 (received for review April 21, 2011) We have used site-directed spin labeling and pulsed electron has prevented the development of a reliable structural model for paramagnetic resonance to resolve a controversy concerning the any of these complexes. A major unresolved question concerns structure of the utrophin–actin complex, with implications for the the relative disposition of the tandem CH domains (CH1 and pathophysiology of muscular dystrophy. Utrophin is a homolog of CH2) (9, 10). Crystal structures of the tandem CH domains dystrophin, the defective protein in Duchenne and Becker muscular showed a closed conformation for fimbrin (11) and α-actinin (12), dystrophies, and therapeutic utrophin derivatives are currently but an open conformation for both utrophin (Utr261) (Fig. 1A) being developed. Both proteins have a pair of N-terminal calponin and dystrophin (Dys246) (16). The crystal structure of Utr261 homology (CH) domains that are important for actin binding. suggests that the central helical region connecting CH1 and CH2 Although there is a crystal structure of the utrophin actin-binding is highly flexible. Even for α-actinin, which has a closed crystal domain, electron microscopy of the actin-bound complexes has structure, computational analysis suggests the potential for a high produced two very different structural models, in which the CH do- degree of dynamic flexibility that facilitates actin binding (17). -

Cyclin D1/Cyclin-Dependent Kinase 4 Interacts with Filamin a and Affects the Migration and Invasion Potential of Breast Cancer Cells
Published OnlineFirst February 28, 2010; DOI: 10.1158/0008-5472.CAN-08-1108 Tumor and Stem Cell Biology Cancer Research Cyclin D1/Cyclin-Dependent Kinase 4 Interacts with Filamin A and Affects the Migration and Invasion Potential of Breast Cancer Cells Zhijiu Zhong, Wen-Shuz Yeow, Chunhua Zou, Richard Wassell, Chenguang Wang, Richard G. Pestell, Judy N. Quong, and Andrew A. Quong Abstract Cyclin D1 belongs to a family of proteins that regulate progression through the G1-S phase of the cell cycle by binding to cyclin-dependent kinase (cdk)-4 to phosphorylate the retinoblastoma protein and release E2F transcription factors for progression through cell cycle. Several cancers, including breast, colon, and prostate, overexpress the cyclin D1 gene. However, the correlation of cyclin D1 overexpression with E2F target gene regulation or of cdk-dependent cyclin D1 activity with tumor development has not been identified. This suggests that the role of cyclin D1 in oncogenesis may be independent of its function as a cell cycle regulator. One such function is the role of cyclin D1 in cell adhesion and motility. Filamin A (FLNa), a member of the actin-binding filamin protein family, regulates signaling events involved in cell motility and invasion. FLNa has also been associated with a variety of cancers including lung cancer, prostate cancer, melanoma, human bladder cancer, and neuroblastoma. We hypothesized that elevated cyclin D1 facilitates motility in the invasive MDA-MB-231 breast cancer cell line. We show that MDA-MB-231 motility is affected by disturbing cyclin D1 levels or cyclin D1-cdk4/6 kinase activity. -

Lectin-Affinity Chromatography Brain Glycoproteomics and Alzheimer
50 DOI 10.1002/prca.201000070 Proteomics Clin. Appl. 2011, 5, 50–56 REVIEW Lectin-affinity chromatography brain glycoproteomics and Alzheimer disease: Insights into protein alterations consistent with the pathology and progression of this dementing disorder D. Allan Butterfield and Joshua B. Owen Department of Chemistry, Center of Membrane Sciences, and Sanders-Brown Center on Aging, University of Kentucky, Lexington KY, USA Alzheimer disease (AD) is a neurodegenerative disorder characterized pathologically by the Received: July 7, 2010 accumulation of senile plaques and neurofibrillary tangles, and both these pathological Revised: August 4, 2010 hallmarks of AD are extensively modified by glycosylation. Mounting evidence shows that Accepted: August 10, 2010 alterations in glycosylation patterns influence the pathogenesis and progression of AD, but the vast number of glycan motifs and potential glycosylation sites of glycoproteins has made the field of glycobiology difficult. However, the advent of glycoproteomics has produced major strides in glycoprotein identification and glycosylation site mapping. The use of lectins, proteins that have strong affinity for specific carbohydrate epitopes, to enrich glycoprotein fractions coupled with modern MS, have yielded techniques to elucidate the glycoproteome in AD. Proteomic studies have identified brain proteins in AD and arguably the earliest form of AD, mild cognitive impairment, with altered affinity for Concanavalin-A and wheat germ agglutinin lectins that are consistent with the pathology and progression of this disorder. This is a relatively nascent field of proteomics research in brain, so future studies of lectin-based brain protein separations may lead to additional insights into AD pathogenesis and progression. Keywords: Alzheimers disease (AD) / Lectin-chromatography / Mild cognitive impairment (MCI) / MS / Synaptic alterations 1 Introduction ized pathologically by the accumulation of senile plaques (SPs) and neurofibrillary tangles (NFTs). -

Targeted Genes and Methodology Details for Neuromuscular Genetic Panels
Targeted Genes and Methodology Details for Neuromuscular Genetic Panels Reference transcripts based on build GRCh37 (hg19) interrogated by Neuromuscular Genetic Panels Next-generation sequencing (NGS) and/or Sanger sequencing is performed Motor Neuron Disease Panel to test for the presence of a mutation in these genes. Gene GenBank Accession Number Regions of homology, high GC-rich content, and repetitive sequences may ALS2 NM_020919 not provide accurate sequence. Therefore, all reported alterations detected ANG NM_001145 by NGS are confirmed by an independent reference method based on laboratory developed criteria. However, this does not rule out the possibility CHMP2B NM_014043 of a false-negative result in these regions. ERBB4 NM_005235 Sanger sequencing is used to confirm alterations detected by NGS when FIG4 NM_014845 appropriate.(Unpublished Mayo method) FUS NM_004960 HNRNPA1 NM_031157 OPTN NM_021980 PFN1 NM_005022 SETX NM_015046 SIGMAR1 NM_005866 SOD1 NM_000454 SQSTM1 NM_003900 TARDBP NM_007375 UBQLN2 NM_013444 VAPB NM_004738 VCP NM_007126 ©2018 Mayo Foundation for Medical Education and Research Page 1 of 14 MC4091-83rev1018 Muscular Dystrophy Panel Muscular Dystrophy Panel Gene GenBank Accession Number Gene GenBank Accession Number ACTA1 NM_001100 LMNA NM_170707 ANO5 NM_213599 LPIN1 NM_145693 B3GALNT2 NM_152490 MATR3 NM_199189 B4GAT1 NM_006876 MYH2 NM_017534 BAG3 NM_004281 MYH7 NM_000257 BIN1 NM_139343 MYOT NM_006790 BVES NM_007073 NEB NM_004543 CAPN3 NM_000070 PLEC NM_000445 CAV3 NM_033337 POMGNT1 NM_017739 CAVIN1 NM_012232 POMGNT2 -

Biomarkers in Tbi and Abi: Present, Future, and Going Nowhere
BIOMARKERS IN TBI AND ABI: PRESENT, FUTURE, AND GOING NOWHERE GABRIEL NEWMAN, PH.D., DIRECTOR: THE NEUROSCIENCE TEAM, TOWSON Disclosures: • No ties to drug companies, labs, or medical equipment companies • Studies being conducted by author at present are in area of Neuromoduation as an intervention in Autism, and Photobiomodulation as an intervention in TBI and ABI, not specifically in reliability of biomarkers for these conditions • Author of presentation is primarily a clinician in practice (70%), not researcher (30%) Lab assays: 1. MEG3 and Interluekin: Drops in MEG3 and rise in interleukin-1β (IL-1β), IL-6, and IL-8 correlate with poor prognosis in TBI. Thought to be specific to TBI, but research will tell… (Shao et al, May 2019, Eur Rev Med Pharmacol Sci). High likelihood for good use in future. Relatively easy to order. 2. S100β: calcium binding protein found in astrocytes, responsible for regulating intracellular levels of calcium; not brain specific, thus it shows up in injury not involving TBI. Must be considered along with trauma history. Also, must be taken an hour after concussion. Problem: Hard to obtain because of time limit. – probably going nowhere (Linda Papa, MD, MSc., Sports Med Arthrosc. 2016 Sep; 24(3): 108–115) 3. Glial Fibrillary Acid Protein (GFAP): Glial Fibrillary Acidic Protein (GFAP) is a promising, brain-specific glial-derived biomarker for MTBI in adults and children. GFAP is released in highly increased amounts into blood serum within an hour of an mTBI injury, and can remain elevated for several days after injury. More feasible, given extended detectability. Papa L, Mittal MK, Ramirez J, et al.; . -

Supplementary Figures
Mena regulates the LINC complex to control actin–nuclear lamina associations, trans-nuclear membrane signalling and cancer gene expression Frederic Li Mow Chee!, Bruno Beernaert!, Alexander Loftus!, Yatendra Kumar", Billie G. C. Griffith!, Jimi C. Wills!, Ann P. Wheeler#, J. Douglas Armstrong$, Maddy Parsons%, Irene M. Leigh,(, Charlotte M. Proby&, Alex von Kriegsheim!, Wendy A. Bickmore", Margaret C. Frame,* & Adam Byron,* Supplementary Information Supplementary Figure 1 Supplementary Figure 2 Supplementary Figure 3 Supplementary Table 1 Supplementary Table 2 Supplementary Table 3 Supplementary Table 4 !Cancer Research UK Edinburgh Centre, Institute of Genetics and Cancer, University of Edinburgh, Edinburgh EH< =XR, UK. "MRC Human Genetics Unit, Institute of Genetics and Cancer, University of Edinburgh, Edinburgh EH< =XU, UK. #Advanced Imaging Resource, Institute of Genetics and Cancer, University of Edinburgh, Edinburgh EH< =XU, UK. $Simons Initiative for the Developing Brain, School of Informatics, University of Edinburgh, Edinburgh EHH IYL, UK. %Randall Centre for Cell and Molecular Biophysics, King’s College London, London SEM MUL, UK. &Division of Molecular and Clinical Medicine, School of Medicine, University of Dundee, Dundee DD <HN, UK. 'Institute of Dentistry, Barts and the London School of Medicine and Dentistry, Queen Mary University of London, London EM =AT, UK. *email: [email protected] or [email protected] 1 a cSCC IAC correlation b cSCC IAC pathways c Core adhesome network ENAH −log10(q) MACF1 CSRP1 Met1 Met4 0 5 10 + + CORO2A Integrin signalling + CFL1 pathway PRNP ILK + HSPB1 PALLD PPFIA1 TES RDX Cytoskeletal regulation + VASP + + ARPC2 by Rho GTPase PPP2CA + Met1 + LASP1 MYH9 + VIM TUBA4A Huntington ITGA3 + disease ITGB4 VCL CAV1 ACTB ROCK1 KTN1 FLNA+ CALR DNA FBLIM1 CORO1B RAC1 + replication +ACTN1 ITGA6 + Met4 ITGAV Parkinson ITGB1 disease Actin cytoskel. -

The Impact of Endogenous Annexin A1 on Glucocorticoid Control of Infl Ammatory Arthritis
Basic and translational research Ann Rheum Dis: first published as 10.1136/annrheumdis-2011-201180 on 5 May 2012. Downloaded from EXTENDED REPORT The impact of endogenous annexin A1 on glucocorticoid control of inß ammatory arthritis Hetal B Patel,1 Kristin N Kornerup,1 AndreÕ LF Sampaio,1 Fulvio DÕAcquisto,1 Michael P Seed,1 Ana Paula Girol,2 Mohini Gray,3 Costantino Pitzalis,1 Sonia M Oliani,2 Mauro Perretti1 ▶ Additional (Þ gures and tables) ABSTRACT Annexin A1 (AnxA1) is an effector of resolution.4 are published online only. To view Objectives To establish the role and effect of Highly expressed in immune cells (eg, polymorpho- these Þ les please visit the journal nuclear cells and macrophages), this protein is exter- online (http://ard.bmj.com/ glucocorticoids and the endogenous annexin A1 (AnxA1) content/early/recent). pathway in inß ammatory arthritis. nalised to exert paracrine and juxtacrine effects, the vast majority of which are mediated by the formyl- 1William Harvey Research Methods Ankle joint mRNA and protein expression Institute, Barts and The London of AnxA1 and its receptors were analysed in peptide receptor type 2 (FPR2/ALX ([Lipoxin A4 School of Medicine, London UK naive and arthritic mice by real-time PCR and receptor]) or FPR2, in rodents).5 Intriguingly, FPR2/ 2Department of Biology; 6 immunohistochemistry. Inß ammatory arthritis was ALX is also the lipoxin A4 receptor indicating the Instituto de Bioci•ncias, Letras +/+ existence of important – yet not fully appreci- e Ci•ncias Exatas (IBILCE), S‹o induced with the K/BxN arthritogenic serum in AnxA1 −/− ated – networks in resolution.7 Paulo State University, S‹o JosŽ and AnxA1 mice; in some experiments, animals Another receptor do Rio Preto, Brazil were treated with dexamethasone (Dex) or with human is also advocated to mediate the effects of AnxA1, 3Medical Research Council recombinant AnxA1 or a protease-resistant mutant the formyl-peptide receptor type 1 or FPR1 (FPR1 Centre for Inß ammation, (termed SuperAnxA1). -

Genetic Variation Screening of TNNT2 Gene in a Cohort of Patients with Hypertrophic and Dilated Cardiomyopathy
Physiol. Res. 61: 169-175, 2012 https://doi.org/10.33549/physiolres.932157 Genetic Variation Screening of TNNT2 Gene in a Cohort of Patients With Hypertrophic and Dilated Cardiomyopathy M. JÁCHYMOVÁ1, A. MURAVSKÁ1, T. PALEČEK2, P. KUCHYNKA2, H. ŘEHÁKOVÁ1, S. MAGAGE2, A. KRÁL2, T. ZIMA1, K. HORKÝ2, A. LINHART2 1Institute of Clinical Chemistry and Laboratory Diagnostics, First Faculty of Medicine and General University Hospital, Charles University, Prague, Czech Republic, 2Second Department of Internal Medicine – Clinical Department of Cardiology and Angiology, First Faculty of Medicine and General University Hospital, Charles University, Prague, Czech Republic Received February 1, 2011 Accepted October 17, 2011 On-line January 31, 2012 Summary Introduction Mutations in troponin T (TNNT2) gene represent the important part of currently identified disease-causing mutations in Cardiomyopathies are generally defined as hypertrophic (HCM) and dilated (DCM) cardiomyopathy. The aim myocardial disorders in which the heart muscle is of this study was to analyze TNNT2 gene exons in patients with structurally and functionally abnormal, in the absence of HCM and DCM diagnosis to improve diagnostic and genetic coronary artery disease, hypertension, valvular disease consultancy in affected families. All 15 exons and their flanking and congenital heart disease sufficient to cause the regions of the TNNT2 gene were analyzed by DNA sequence observed myocardial abnormality (Elliott et al. 2008). analysis in 174 patients with HCM and DCM diagnosis. We According to the morphological and functional phenotype identified genetic variations in TNNT2 exon regions in 56 patients the diagnosis of hypertrophic and dilated cardiomyopathy and genetic variations in TNNT2 intron regions in 164 patients. -

Analysis of Gene Expression Data for Gene Ontology
ANALYSIS OF GENE EXPRESSION DATA FOR GENE ONTOLOGY BASED PROTEIN FUNCTION PREDICTION A Thesis Presented to The Graduate Faculty of The University of Akron In Partial Fulfillment of the Requirements for the Degree Master of Science Robert Daniel Macholan May 2011 ANALYSIS OF GENE EXPRESSION DATA FOR GENE ONTOLOGY BASED PROTEIN FUNCTION PREDICTION Robert Daniel Macholan Thesis Approved: Accepted: _______________________________ _______________________________ Advisor Department Chair Dr. Zhong-Hui Duan Dr. Chien-Chung Chan _______________________________ _______________________________ Committee Member Dean of the College Dr. Chien-Chung Chan Dr. Chand K. Midha _______________________________ _______________________________ Committee Member Dean of the Graduate School Dr. Yingcai Xiao Dr. George R. Newkome _______________________________ Date ii ABSTRACT A tremendous increase in genomic data has encouraged biologists to turn to bioinformatics in order to assist in its interpretation and processing. One of the present challenges that need to be overcome in order to understand this data more completely is the development of a reliable method to accurately predict the function of a protein from its genomic information. This study focuses on developing an effective algorithm for protein function prediction. The algorithm is based on proteins that have similar expression patterns. The similarity of the expression data is determined using a novel measure, the slope matrix. The slope matrix introduces a normalized method for the comparison of expression levels throughout a proteome. The algorithm is tested using real microarray gene expression data. Their functions are characterized using gene ontology annotations. The results of the case study indicate the protein function prediction algorithm developed is comparable to the prediction algorithms that are based on the annotations of homologous proteins. -

Annexina7 and CAP1 Are Associated with Regulating Tumor Cell Adhesion Molecule Expression and Biological Behavior
AnnexinA7 and CAP1 are associated with regulating tumor cell adhesion molecule expression and biological behavior Type Research paper Keywords hepatocellular carcinoma, adhesion molecule, AnnexinA7, CAP1, Biological behaviors Abstract Introduction To find out the correlations between the effects of down-regulating AnnexinA7 and CAP1 gene on cell adhesion factors and the biological behavior of Hca-p cells cells, and finally infer the relationship between the two genes. Material and methods Western blot ,qRT-PCR,immunocytochemistry, CCK8 cell proliferation, flow cytometry, lymph node adhesion and transwell chamber assay testing . Results AnnexinA7 and CAP1 were consistent with the regulation of the expression of the adhesion molecules such as FAK, Src, Paxillin and E-cadherin. At the mRNA and protein levels, the expression of FAK, Src and Paxillin were increased with down-regulated AnnexinA7 and CAP1 genes, while E-cadherin was down-regulated in the change of these two genes. And the low expression of AnnexinA7 could affect the expression of CAP1 in mRNA and protein levels. otherwise, the localization of AnnexinA7 and CAP1 in hepatocellular carcinoma cells was also the same. After down-regulating the expression of CAP1, the functions of proliferation, lymph node adhesion and invasion were increased and earlyPreprint apoptotic ability was decreased in Hca-P cells. Conclusions AnnexinA7 and CAP1 could control the expression of these adhesion molecules in the same trend. We speculated they may be co-localization. And AnnexinA7 gene may be related to the molecular mechanism of CAP1 gene, which is likely to have a consistent effect on cell adhesion molecules and be closely related to the biological behaviors of Hca-P cells.AnnexinA7 and CAP1 may play an inhibitory role in lymph node metastasis to provide a reliable basis for the early identification of lymphatic metastasis in hepatocellular carcinoma. -

Impaired Immune Surveillance Accelerates Accumulation of Senescent Cells and Aging
ARTICLE https://doi.org/10.1038/s41467-018-07825-3 OPEN Impaired immune surveillance accelerates accumulation of senescent cells and aging Yossi Ovadya1, Tomer Landsberger2, Hanna Leins3,4, Ezra Vadai1, Hilah Gal1, Anat Biran1, Reut Yosef1, Adi Sagiv1, Amit Agrawal1, Alon Shapira1, Joseph Windheim1, Michael Tsoory5, Reinhold Schirmbeck4, Ido Amit 2, Hartmut Geiger3,6 & Valery Krizhanovsky 1 Cellular senescence is a stress response that imposes stable cell-cycle arrest in damaged 1234567890():,; cells, preventing their propagation in tissues. However, senescent cells accumulate in tissues in advanced age, where they might promote tissue degeneration and malignant transfor- mation. The extent of immune-system involvement in regulating age-related accumulation of senescent cells, and its consequences, are unknown. Here we show that Prf1−/− mice with impaired cell cytotoxicity exhibit both higher senescent-cell tissue burden and chronic inflammation. They suffer from multiple age-related disorders and lower survival. Strikingly, pharmacological elimination of senescent-cells by ABT-737 partially alleviates accelerated aging phenotype in these mice. In LMNA+/G609G progeroid mice, impaired cell cytotoxicity further promotes senescent-cell accumulation and shortens lifespan. ABT-737 administration during the second half of life of these progeroid mice abrogates senescence signature and increases median survival. Our findings shed new light on mechanisms governing senescent- cell presence in aging, and could motivate new strategies for regenerative medicine. 1 Department of Molecular Cell Biology, The Weizmann Institute of Science, 76100 Rehovot, Israel. 2 Department of Immunology, The Weizmann Institute of Science, 76100 Rehovot, Israel. 3 Institute of Molecular Medicine, Stem Cell and Aging, Ulm University, Ulm 89081, Germany. -

Keratins and Plakin Family Cytolinker Proteins Control the Length Of
RESEARCH ARTICLE Keratins and plakin family cytolinker proteins control the length of epithelial microridge protrusions Yasuko Inaba*, Vasudha Chauhan, Aaron Paul van Loon, Lamia Saiyara Choudhury, Alvaro Sagasti* Molecular, Cell and Developmental Biology Department and Molecular Biology Institute, University of California, Los Angeles, Los Angeles, United States Abstract Actin filaments and microtubules create diverse cellular protrusions, but intermediate filaments, the strongest and most stable cytoskeletal elements, are not known to directly participate in the formation of protrusions. Here we show that keratin intermediate filaments directly regulate the morphogenesis of microridges, elongated protrusions arranged in elaborate maze-like patterns on the surface of mucosal epithelial cells. We found that microridges on zebrafish skin cells contained both actin and keratin filaments. Keratin filaments stabilized microridges, and overexpressing keratins lengthened them. Envoplakin and periplakin, plakin family cytolinkers that bind F-actin and keratins, localized to microridges, and were required for their morphogenesis. Strikingly, plakin protein levels directly dictate microridge length. An actin-binding domain of periplakin was required to initiate microridge morphogenesis, whereas periplakin-keratin binding was required to elongate microridges. These findings separate microridge morphogenesis into distinct steps, expand our understanding of intermediate filament functions, and identify microridges as protrusions that integrate actin and intermediate filaments. *For correspondence: [email protected] (YI); Introduction [email protected] (AS) Cytoskeletal filaments are scaffolds for membrane protrusions that create a vast diversity of cell shapes. The three major classes of cytoskeletal elements—microtubules, actin filaments, and inter- Competing interests: The mediate filaments (IFs)—each have distinct mechanical and biochemical properties and associate authors declare that no with different regulatory proteins, suiting them to different functions.