Rna Ligation by Hammerhead Ribozymes and Dnazyme In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Hammerhead Ribozymes Against Virus and Viroid Rnas
Hammerhead Ribozymes Against Virus and Viroid RNAs Alberto Carbonell, Ricardo Flores, and Selma Gago Contents 1 A Historical Overview: Hammerhead Ribozymes in Their Natural Context ................................................................... 412 2 Manipulating Cis-Acting Hammerheads to Act in Trans ................................. 414 3 A Critical Issue: Colocalization of Ribozyme and Substrate . .. .. ... .. .. .. .. .. ... .. .. .. .. 416 4 An Unanticipated Participant: Interactions Between Peripheral Loops of Natural Hammerheads Greatly Increase Their Self-Cleavage Activity ........................... 417 5 A New Generation of Trans-Acting Hammerheads Operating In Vitro and In Vivo at Physiological Concentrations of Magnesium . ...... 419 6 Trans-Cleavage In Vitro of Short RNA Substrates by Discontinuous and Extended Hammerheads ........................................... 420 7 Trans-Cleavage In Vitro of a Highly Structured RNA by Discontinuous and Extended Hammerheads ........................................... 421 8 Trans-Cleavage In Vivo of a Viroid RNA by an Extended PLMVd-Derived Hammerhead ........................................... 422 9 Concluding Remarks and Outlooks ........................................................ 424 References ....................................................................................... 425 Abstract The hammerhead ribozyme, a small catalytic motif that promotes self- cleavage of the RNAs in which it is found naturally embedded, can be manipulated to recognize and cleave specifically -

Ribozymes Targeted to the Mitochondria Using the 5S Ribosomal Rna
RIBOZYMES TARGETED TO THE MITOCHONDRIA USING THE 5S RIBOSOMAL RNA By JENNIFER ANN BONGORNO A DISSERTATION PRESENTED TO THE GRADUATE SCHOOL OF THE UNIVERSITY OF FLORIDA IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY UNIVERSITY OF FLORIDA 2005 Copyright 2005 by Jennifer Bongorno To my grandmother, Hazel Traster Miller, whose interest in genealogy sparked my interest in genetics, and without whose mitochondria I would not be here ACKNOWLEDGMENTS I would like to thank all the members of the Lewin lab; especially my mentor, Al Lewin. Al was always there for me with suggestions and keeping me motivated. He and the other members of the lab were like my second family; I would not have had an enjoyable experience without them. Diana Levinson and Elizabeth Bongorno worked with me on the fourth and third mouse transfections respectively. Joe Hartwich and Al Lewin tested some of the ribozymes in vitro and cloned some of the constructs I used. James Thomas also helped with cloning and was an invaluable lab manager. Verline Justilien worked on a related project and was a productive person with whom to bounce ideas back and forth. Lourdes Andino taught me how to use the new phosphorimager for my SYBR Green-stained gels. Alan White was there through it all, like the older brother I never had. Mary Ann Checkley was with me even longer than Alan, since we both came to Florida from Ohio Wesleyan, although she did manage to graduate before me. Jia Liu and Frederic Manfredsson were there when I needed a beer. -

In Vitro Analysis of the Self-Cleaving Satellite RNA of Barley Yellow Dwarf Virus Stanley Livingstone Silver Iowa State University
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 1993 In vitro analysis of the self-cleaving satellite RNA of barley yellow dwarf virus Stanley Livingstone Silver Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Biochemistry Commons, Molecular Biology Commons, and the Plant Pathology Commons Recommended Citation Silver, Stanley Livingstone, "In vitro analysis of the self-cleaving satellite RNA of barley yellow dwarf virus " (1993). Retrospective Theses and Dissertations. 10274. https://lib.dr.iastate.edu/rtd/10274 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. _UMI MICROFILMED 1993 | INFORMATION TO USERS This manuscript has been reproduced from the microfilm master. UMI films the text directly from the original or copy submitted. Thus, some thesis and dissertation copies are in typewriter face, while others may be from any type of computer printer. The quality of this reproduction is dependent upon the quality of the copy submitted. Broken or indistinct print, colored or poor quality illustrations and photographs, print bleedthrough, substandard margins, and improper alignment can adversely affect reproduction. In the unlikely event that the author did not send UMI a complete manuscript and there are missing pages, these will be noted. Also, if unauthorized copyright material had to be removed, a note will indicate the deletion. -

(10) Patent No.: US 9139554 B2
US009139554B2 (12) United States Patent (10) Patent No.: US 9,139,554 B2 Hope et al. (45) Date of Patent: Sep. 22, 2015 (54) AMINO LIPIDS AND METHODS FOR THE 4,438,052 A 3, 1984 Weder et al. DELIVERY OF NUCLECACDS 4,469,863. A 9, 1984 Ts'o et al. 4476,301 A 10, 1984 Imbach et al. 4,515,736 A 5, 1985 Deamer (75) Inventors: Michael J. Hope, Vancouver (CA), Sean 4,522,803 A 6, 1985 Lenk et al. C. Semple, Delta (CA); Jianxin Chen, 4,588,578 A 5/1986 Fountain et al. Vancouver (CA); Thomas D. Madden, 4,598,051 A 7, 1986 Papahadjopoulos et al. Vancouver (CA); Pieter R. Cullis, - 2: A 3. E. SR etCa al Vancouver SCA) Marco A. Ciufolini, 4,839, 175 A ck 6, 1989 Guo et al. 424/450 Vancouver (CA), Barbara Low Shoud 4,845.205 A 7/1989 Huynh Dinh et al. Mui, Vancouver (CA) 4,897,355 A 1/1990 Eppstein et al. 4,957,773. A 9/1990 Spencer et al. (73) Assignees: Tekmira Pharmaceuticals E.- w A 3. : ech et calal. E.niversity of Briticini,British Columbia, 5,023,2435,013,556 A 6/19915/1991 WoodleTullis et al. Vancouver (CA) 5,034,506 A 7/1991 Summerton et al. 5,118,800 A 6/1992 Smith et al. (*) Notice: Subject to any disclaimer, the term of this 5,130,302 A 7/1992 Spielvogel et al. patent is extended or adjusted under 35 3. A RE SRN. tal U.S.C. -

Hepatitis Delta: Virological and Clinical Aspects
Botelho-Souza et al. Virology Journal (2017) 14:177 DOI 10.1186/s12985-017-0845-y REVIEW Open Access Hepatitis delta: virological and clinical aspects Luan Felipo Botelho-Souza1,2,3*, Mariana Pinheiro Alves Vasconcelos4, Alcione de Oliveira dos Santos1,2,3, Juan Miguel Villalobos Salcedo1,2,3 and Deusilene Souza Vieira1,2,3 Abstract There are an estimated 400 million chronic carriers of HBV worldwide; between 15 and 20 million have serological evidence of exposure to HDV. Traditionally, regions with high rates of endemicity are central and northern Africa, the Amazon Basin, eastern Europe and the Mediterranean, the Middle East and parts of Asia. There are two types of HDV/HBV infection which are differentiated by the previous status infection by HBV for the individual. Individuals with acute HBV infection contaminated by HDV is an HDV/HBV co-infection, while individuals with chronic HBV infection contaminated by HDV represent an HDV/HBV super-infection. The appropriate treatment for chronic hepatitis delta is still widely discussed since it does not have an effective drug. Alpha interferon is currently the only licensed therapy for the treatment of chronic hepatitis D. The most widely used drug is pegylated interferon but only approximately 25% of patients maintain a sustained viral response after 1 year of treatment. The best marker of therapeutic success would be the clearance of HBsAg, but this data is rare in clinical practice. Therefore, the best way to predict a sustained virologic response is the maintenance of undetectable HDV RNA levels. Keywords: HDV, Virology, Clinical aspects Background Later it was demonstrated that HBV infection associ- The hepatitis delta or D virus (HDV) was discovered in ated with HDAg (HBV/HDV) did not develop in chim- 1977 by Rizzetto et al. -

Review Chemistry and Biology of Self-Cleaving Ribozymes Randi M
TIBS 1181 No. of Pages 14 Review Chemistry and Biology of Self-Cleaving Ribozymes Randi M. Jimenez,1 Julio A. Polanco,1 and Andrej Lupták1,2,3,* Self-cleaving ribozymes were discovered 30 years ago, but their biological Trends fi distribution and catalytic mechanisms are only beginning to be de ned. Each Self-cleaving ribozymes are distributed ribozyme family is defined by a distinct structure, with unique active sites throughout all branches of life. Cur- accelerating the same transesterification reaction across the families. Biochem- rently, there are nine distinct structural motifs that promote self-scission in ical studies show that general acid-base catalysis is the most common mecha- nature. nism of self-cleavage, but metal ions and metabolites can be used as cofactors. The six self-cleaving ribozymes that Ribozymes have been discovered in highly diverse genomic contexts through- have been investigated mechanistically out nature, from viroids to vertebrates. Their biological roles include self- all appear to use a general acid-base scission during rolling-circle replication of RNA genomes, co-transcriptional mechanism for catalysis. Magnesium, or another divalent metal ion, is largely processing of retrotransposons, and metabolite-dependent gene expression used to stabilize the tertiary structures regulation in bacteria. Other examples, including highly conserved mammalian of these ribozymes. ribozymes, suggest that many new biological roles are yet to be discovered. The broad distribution of self-cleaving ribozymes suggests several biological Guiding Principles for Ribozyme Exploration roles. The known functions include RNA processing during rolling-circle Small nucleolytic ribozymes carry out site-specific phosphodiester scission without the need for replication of single-stranded subviral protein chaperones or enzymes. -

VIROLOGY-I Viruses: Structure and Definition Viruses
VIROLOGY-I Viruses: Structure and Definition Viruses • A virus is a small parasite that cannot reproduce by itself. • can direct the cell machinery to produce more viruses. • have either RNA or DNA as their genetic material. The nucleic acid may be single- or double-stranded. History of the Discovery of Viruses • the word “virus” comes from a Latin word simply meaning “slimy fluid” • Adolf Eduard Mayer from Germany, publishing in 1886 on work done in Holland from 1879, showed that the “mosaic disease” of tobacco – or “mozaïkziekte”, as he named it in his paper – could be transmitted to other plants by rubbing a liquid extract, filtered through paper, from an infected plant onto the leaves of a healthy plant. The second virus discovered was what is now known as Foot and mouth disease virus (FMDV) of farm and other animals, in 1898by the German scientists Friedrich Loeffler and Paul Frosch. Again, their “sterile” filtered liquid proved infectious in calves, providing the first proof of viruses infecting animals In 1898 G Sanarelli, working in Uruguay, described the smallpox virus relative and tumour-causing myxoma virus of rabbits as a virus – but on the basis of sterilisation by centrifugation rather than by filtration. A finding that was later to have great importance in veterinary virology was the discovery by Maurice Nicolle and Adil Mustafa (also known as Adil-Bey), in Turkey in 1902, that rinderpest or cattle plague was caused by a virus. This had been for several centuries the worst animal disease known worldwide in terms of mortality: for example, an epizootic or animal epidemic in Africa in the 1890s that had started in what is now Ethiopia in 1887 from cattle imported from Asia, had spread throughout the continent by 1897, and killed 80-90% of the cattle and a large proportion of susceptible wild animals in southern Africa. -

HDV Family of Self-Cleaving Ribozymes
Provided for non-commercial research and educational use only. Not for reproduction, distribution or commercial use. This chapter was originally published in the book Progress in Molecular Biology and Translational Science, Vol. 120, published by Elsevier, and the attached copy is provided by Elsevier for the author's benefit and for the benefit of the author's institution, for non-commercial research and educational use including without limitation use in instruction at your institution, sending it to specific colleagues who know you, and providing a copy to your institution’s administrator. All other uses, reproduction and distribution, including without limitation commercial reprints, selling or licensing copies or access, or posting on open internet sites, your personal or institution’s website or repository, are prohibited. For exceptions, permission may be sought for such use through Elsevier's permissions site at: http://www.elsevier.com/locate/permissionusematerial From: Nathan Riccitelli, Andrej Lupták, HDV Family of Self-Cleaving Ribozymes. In Garrett A. Soukup, editor: Progress in Molecular Biology and Translational Science, Vol. 120, Burlington: Academic Press, 2013, pp. 123-171. ISBN: 978-0-12-381286-5 © Copyright 2013 Elsevier Inc. Academic Press Author's personal copy CHAPTER FOUR HDV Family of Self-Cleaving Ribozymes Nathan Riccitelli*, Andrej Lupták*,†,{ *Department of Chemistry, University of California, Irvine, California, USA †Department of Pharmaceutical Sciences, University of California, Irvine, California, USA { Department of Molecular Biology and Biochemistry, University of California, Irvine, California, USA Contents 1. Introduction 124 2. Hepatitis Delta Virus 124 3. HDV Ribozymes 127 3.1 Ribozyme structure 127 3.2 Ribozyme folding 129 4. -

Multipartite Viruses: Organization, Emergence and Evolution
UNIVERSIDAD AUTÓNOMA DE MADRID FACULTAD DE CIENCIAS DEPARTAMENTO DE BIOLOGÍA MOLECULAR MULTIPARTITE VIRUSES: ORGANIZATION, EMERGENCE AND EVOLUTION TESIS DOCTORAL Adriana Lucía Sanz García Madrid, 2019 MULTIPARTITE VIRUSES Organization, emergence and evolution TESIS DOCTORAL Memoria presentada por Adriana Luc´ıa Sanz Garc´ıa Licenciada en Bioqu´ımica por la Universidad Autonoma´ de Madrid Supervisada por Dra. Susanna Manrubia Cuevas Centro Nacional de Biotecnolog´ıa (CSIC) Memoria presentada para optar al grado de Doctor en Biociencias Moleculares Facultad de Ciencias Departamento de Biolog´ıa Molecular Universidad Autonoma´ de Madrid Madrid, 2019 Tesis doctoral Multipartite viruses: Organization, emergence and evolution, 2019, Madrid, Espana. Memoria presentada por Adriana Luc´ıa-Sanz, licenciada en Bioqumica´ y con un master´ en Biof´ısica en la Universidad Autonoma´ de Madrid para optar al grado de doctor en Biociencias Moleculares del departamento de Biolog´ıa Molecular en la facultad de Ciencias de la Universidad Autonoma´ de Madrid Supervisora de tesis: Dr. Susanna Manrubia Cuevas. Investigadora Cient´ıfica en el Centro Nacional de Biotecnolog´ıa (CSIC), C/ Darwin 3, 28049 Madrid, Espana. to the reader CONTENTS Acknowledgments xi Resumen xiii Abstract xv Introduction xvii I.1 What is a virus? xvii I.2 What is a multipartite virus? xix I.3 The multipartite lifecycle xx I.4 Overview of this thesis xxv PART I OBJECTIVES PART II METHODOLOGY 0.5 Database management for constructing the multipartite and segmented datasets 3 0.6 Analytical -

Subdomain Cryo-EM Structure of Nodaviral Replication Protein a Crown Complex Provides Mechanistic Insights Into RNA Genome Replication
Subdomain cryo-EM structure of nodaviral replication protein A crown complex provides mechanistic insights into RNA genome replication Nuruddin Unchwaniwalaa,b,c,1, Hong Zhana,b,c,1, Janice Penningtonb,c, Mark Horswilla,b,c, Johan A. den Boona,b,c, and Paul Ahlquista,b,c,d,2 aJohn and Jeanne Rowe Center for Research in Virology, Morgridge Institute for Research, Madison, WI 53715; bInstitute for Molecular Virology, University of Wisconsin–Madison, Madison, WI 53706; cMcArdle Laboratory for Cancer Research, University of Wisconsin–Madison, Madison, WI 53706; and dHoward Hughes Medical Institute, University of Wisconsin–Madison, Madison, WI 53706 Contributed by Paul Ahlquist, June 9, 2020 (sent for review April 2, 2020; reviewed by Ralf Bartenschlager and Richard J. Kuhn) For positive-strand RNA [(+)RNA] viruses, the major target for an- templates, and provide the enzymatic functions for viral RNA syn- tiviral therapies is genomic RNA replication, which occurs at poorly thesis and often 5′ m7G capping. understood membrane-bound viral RNA replication complexes. Re- Classic electron tomography has greatly illuminated the dra- cent cryoelectron microscopy (cryo-EM) of nodavirus RNA replication matic membrane rearrangements associated with such (+)RNA complexes revealed that the viral double-stranded RNA replication virus replication complexes (4–7). However, due to multiple template is coiled inside a 30- to 90-nm invagination of the outer limitations of traditional electron microscopy (EM) sample prep- mitochondrial membrane, whose necked aperture to the cytoplasm aration and visualization by heavy metal staining, such studies lack “ ” is gated by a 12-fold symmetric, 35-nm diameter crown complex information on the organization of viral proteins and RNAs at that contains multifunctional viral RNA replication protein A. -

Hepatitis Delta Virus: Replication Strategy and Upcoming Therapeutic Options for a Neglected Human Pathogen
viruses Review Hepatitis Delta Virus: Replication Strategy and Upcoming Therapeutic Options for a Neglected Human Pathogen Florian A. Lempp 1,2 and Stephan Urban 1,2,* 1 Department of Infectious Diseases, Molecular Virology, Heidelberg University, Im Neuenheimer Feld 345, 69120 Heidelberg, Germany; [email protected] 2 German Centre for Infection Research (DZIF), Partner Site Heidelberg, 69120 Heidelberg, Germany * Correspondence: [email protected]; Tel.: +49-6221-564902 Academic Editors: Ulrike Protzer and Michael Nassal Received: 12 June 2017; Accepted: 29 June 2017; Published: 4 July 2017 Abstract: The human Hepatitis Delta Virus (HDV) is unique among all viral pathogens. Encoding only one protein (Hepatitis Delta Antigen; HDAg) within its viroid-like self-complementary RNA, HDV constitutes the smallest known virus in the animal kingdom. To disseminate in its host, HDV depends on a helper virus, the human Hepatitis B virus (HBV), which provides the envelope proteins required for HDV assembly. HDV affects an estimated 15–20 million out of the 240 million chronic HBV-carriers and disperses unequally in disparate geographical regions of the world. The disease it causes (chronic Hepatitis D) presents as the most severe form of viral hepatitis, leading to accelerated progression of liver dysfunction including cirrhosis and hepatocellular carcinoma and a high mortality rate. The lack of approved drugs interfering with specific steps of HDV replication poses a high burden for gaining insights into the molecular biology of the virus and, consequently, the development of specific novel medications that resiliently control HDV replication or, in the best case, functionally cure HDV infection or HBV/HDV co-infection. -
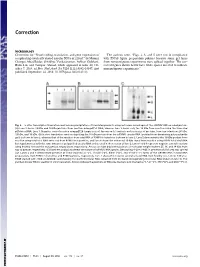
Novel Coding, Translation, and Gene Expression of a Replicating Covalently Closed Circular RNA of 220 Nt
Correction MICROBIOLOGY Correction for “Novel coding, translation, and gene expression of The authors note, “Figs. 2, 3, and 5 were not in compliance a replicating covalently closed circular RNA of 220 nt,” by Mounir with PNAS figure preparation policies because some gel lanes Georges AbouHaidar, Srividhya Venkataraman, Ashkan Golshani, from noncontiguous experiments were spliced together. The cor- Bolin Liu, and Tauqeer Ahmad, which appeared in issue 40, Oc- rected figures shown below have white spaces inserted to indicate tober 7, 2014, of Proc Natl Acad Sci USA (111:14542–14547; first noncontiguous experiments.” published September 24, 2014; 10.1073/pnas.1402814111). Fig. 2. In vitro transcription/translation and immunoprecipitation of translated products using antiserum raised against the scRYMV ORF-encoded protein. (A) Lane 2 shows 19-kDa and 16-kDa proteins from reaction using pET52 DNA, whereas lane 3 shows only the 19 kDa from reaction using the truncated pETdimer DNA. Lane 1: Negative control reaction using pET29 (empty vector). Arrows on left indicate molecular size of proteins, from top to bottom: 36 kDa, 19 kDa, and 16 kDa. (B) In vitro translation reaction depicting the 16-kDa protein from the scRYMV circular RNA (purified from denaturing polyacrylamide gels) is shown in lane 3, whereas that of the reaction from total RNA of RYMV-infected rice is shown in lane 2. Lane 5 demonstrates the 16-kDa product from reaction using total viral RNA extracted from RYMV virus particles, and lane 4 shows the enhanced 16-kDa signal from reaction using RYMV total viral RNA but supplemented with the same amount of gel-purified circular RNA as that used in the reaction of lane 3.