An Advanced System of the Mitochondrial Processing Peptidase and Core Protein Family in Trypanosoma Brucei and Multiple Origins of the Core I Subunit in Eukaryotes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
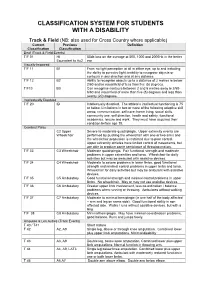
Disability Classification System
CLASSIFICATION SYSTEM FOR STUDENTS WITH A DISABILITY Track & Field (NB: also used for Cross Country where applicable) Current Previous Definition Classification Classification Deaf (Track & Field Events) T/F 01 HI 55db loss on the average at 500, 1000 and 2000Hz in the better Equivalent to Au2 ear Visually Impaired T/F 11 B1 From no light perception at all in either eye, up to and including the ability to perceive light; inability to recognise objects or contours in any direction and at any distance. T/F 12 B2 Ability to recognise objects up to a distance of 2 metres ie below 2/60 and/or visual field of less than five (5) degrees. T/F13 B3 Can recognise contours between 2 and 6 metres away ie 2/60- 6/60 and visual field of more than five (5) degrees and less than twenty (20) degrees. Intellectually Disabled T/F 20 ID Intellectually disabled. The athlete’s intellectual functioning is 75 or below. Limitations in two or more of the following adaptive skill areas; communication, self-care; home living, social skills, community use, self direction, health and safety, functional academics, leisure and work. They must have acquired their condition before age 18. Cerebral Palsy C2 Upper Severe to moderate quadriplegia. Upper extremity events are Wheelchair performed by pushing the wheelchair with one or two arms and the wheelchair propulsion is restricted due to poor control. Upper extremity athletes have limited control of movements, but are able to produce some semblance of throwing motion. T/F 33 C3 Wheelchair Moderate quadriplegia. Fair functional strength and moderate problems in upper extremities and torso. -

MIPS IV Instruction Set
MIPS IV Instruction Set Revision 3.2 September, 1995 Charles Price MIPS Technologies, Inc. All Right Reserved RESTRICTED RIGHTS LEGEND Use, duplication, or disclosure of the technical data contained in this document by the Government is subject to restrictions as set forth in subdivision (c) (1) (ii) of the Rights in Technical Data and Computer Software clause at DFARS 52.227-7013 and / or in similar or successor clauses in the FAR, or in the DOD or NASA FAR Supplement. Unpublished rights reserved under the Copyright Laws of the United States. Contractor / manufacturer is MIPS Technologies, Inc., 2011 N. Shoreline Blvd., Mountain View, CA 94039-7311. R2000, R3000, R6000, R4000, R4400, R4200, R8000, R4300 and R10000 are trademarks of MIPS Technologies, Inc. MIPS and R3000 are registered trademarks of MIPS Technologies, Inc. The information in this document is preliminary and subject to change without notice. MIPS Technologies, Inc. (MTI) reserves the right to change any portion of the product described herein to improve function or design. MTI does not assume liability arising out of the application or use of any product or circuit described herein. Information on MIPS products is available electronically: (a) Through the World Wide Web. Point your WWW client to: http://www.mips.com (b) Through ftp from the internet site “sgigate.sgi.com”. Login as “ftp” or “anonymous” and then cd to the directory “pub/doc”. (c) Through an automated FAX service: Inside the USA toll free: (800) 446-6477 (800-IGO-MIPS) Outside the USA: (415) 688-4321 (call from a FAX machine) MIPS Technologies, Inc. -

2019 NFCA Texas High School Leadoff Classic Main Bracket Results
2019 NFCA Texas High School Leadoff Classic Main Bracket Results Bryan College Station BHS CSHS 1 Bryan College Station 17 Thu 3pm Thu 3pm Cedar Creek BHS CSHS EP Eastlake SA Brandeis 49 Brandeis College Station 57 Cy Woods 1 BHS Thu 5pm Thu 5pm CSHS 2 18 Thu 1pm Brandeis Robinson Thu 1pm EP Montwood BHS CSHS Robinson 11 Huntsville 3 89 Klein Splendora 93 Splendora 9 BHS Fri 2pm Fri 2pm CSHS 3 Fredericksburg Splendora 19 Thu 9am Thu 9am Fredericksburg 6 VET1 VET2 Belton 6 Grapevine 50 58 Richmond Foster 11 BHS Thu 5pm Klein Splendora Thu 5pm CSHS 4 20 Thu 11am Klein Richmond Foster Thu 11am Klein 11 BHS CSHS Rockwall 2 SA Southwest 9 97 SA Southwest Cedar Ridge 99 Cy Ranch 7 VET1 Fri 4pm Fri 4pm CP3 5 Southwest Cy Ranch 21 Thu 11am Thu 1pm Temple 5 VET1 CP3 Plano East 5 Clear Springs 2 51 Southwest Alvin 59 Alvin VET1 Thu 3pm Thu 5pm CP3 6 22 Thu 9am Vandegrift Alvin Thu 3pm Vandegrift 5 BHS CSHS Lufkin San Marcos 5 90 94 Flower Mound 0 VET2 Fri 12pm Southwest Cedar Ridge Fri 12pm CP4 7 San Marcos Cedar Ridge 23 Thu 9am Thu 1pm Magnolia West 3 VET2 CHAMPIONSHIP CP4 RR Cedar Ridge 12 McKinney Boyd 3 52 Game 192 60 SA Johnson VET2 Thu 3pm San Marcos BHS Cedar Ridge Thu 5 pm CP4 8 4:00 PM 24 Thu 11am M. Boyd BHS 5 10 BHS Johnson Thu 3pm Manvel 0 129 Southwest vs. Cedar Ridge 130 Kingwood Park Bellaire 3 Sat 10am Sat 8am Deer Park VET4 BRAC-BB 9 Cedar Park Deer Park 25 Thu 11am Thu 1pm Cedar Park 5 VET4 BRAC-BB Leander Clements 13 53 Cedar Park MacArthur 61 SA MacArthur VET4 Thu 3pm Thu 5pm BRAC-BB 10 26 Thu 9am Clements MacArthur Thu 3pm Waco University 0 BHS CSHS Tomball Memorial Cy Fair 4 91 Friendswood Woodlands 95 Woodlands 2 VET5 Fri 8am Fri 8am BRAC-YS 11 San Benito Woodlands 27 Thu 11am Thu 1pm San Benito 10 VET5 BRAC-YS SA Holmes 1 Friendswood 10 54 62 Santa Fe VET5 Thu 3pm Friendswood Woodlands Thu 5pm BRAC-YS 12 28 Thu 9am Friendswood Santa Fe Thu 3pm Henderson 0 VET1 VET2 Lake Travis Ridge Point 16 98 100 B. -

Classification Made Easy Class 1
Classification Made Easy Class 1 (CP1) The most severely disabled athletes belong to this classification. These athletes are dependent on a power wheelchair or assistance for mobility. They have severe limitation in both the arms and the legs and have very poor trunk control. Sports Available: • Race Runner (RR1) – using the Race Runner frame to run, track events include 100m, 200m and 400m. • Boccia o Boccia Class 1 (BC1) – players who fit into this category can throw the ball onto the court or a CP2 Lower who chooses to push the ball with the foot. Each BC1 athlete has a sport assistant on court with them. o Boccia Class 3 (BC3) – players who fit into this category cannot throw the ball onto the court and have no sustained grasp or release action. They will use a “chute” or “ramp” with the help from their sport assistant to propel the ball. They may use head or arm pointers to hold and release the ball. Players with a impairment of a non cerebral origin, severely affecting all four limbs, are included in this class. Class 2 (CP2) These athletes have poor strength or control all limbs but are able to propel a wheelchair. Some Class 2 athletes can walk but can never run functionally. The class 2 athletes can throw a ball but demonstrates poor grasp and release. Sports Available: • Race Runner (RR2) - using the Race Runner frame to run, track events include 100m, 200m and 400m. • Boccia o Boccia Class 2 (BC2) – players can throw the ball into the court consistently and do not need on court assistance. -

The US Livestock Industry
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 1983 The SU livestock industry: an evaluation of the adequacy and relevance of three models of consumer and producer behavior Stephen Stanley Steyn Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Agricultural and Resource Economics Commons, and the Agricultural Economics Commons Recommended Citation Steyn, Stephen Stanley, "The SU livestock industry: an evaluation of the adequacy and relevance of three models of consumer and producer behavior " (1983). Retrospective Theses and Dissertations. 7653. https://lib.dr.iastate.edu/rtd/7653 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. INFORMATION TO USERS This reproduction was made from a copy of a document sent to us for microfilming. While the most advanced technology has been used to photograph and reproduce this document, the quality of the reproduction is heavily dependent upon the quality of the material submitted. The following explanation of techniques is provided to help clarify markings or notations which may appear on this reproduction. 1. The sign or "target" for pages apparently lacking from the document photographed is "Missing Page(s)". If it was possible to obtain the missing page(s) or section, they are spliced into the film along with adjacent pages. This may have necessitated cutting through an image and duplicating adjacent pages to assure complete continuity. -

Technology Enhancement and Ethics in the Paralympic Games
UNIVERSITY OF PELOPONNESE FACULTY OF HUMAN MOVEMENT AND QUALITY OF LIFE SCIENCES DEPARTMENT OF SPORTS ORGANIZATION AND MANAGEMENT MASTER’S THESIS “OLYMPIC STUDIES, OLYMPIC EDUCATION, ORGANIZATION AND MANAGEMENT OF OLYMPIC EVENTS” Technology Enhancement and Ethics In the Paralympic Games Stavroula Bourna Sparta 2016 i TECHNOLOGY ENHANCEMENT AND ETHICS IN THE PARALYMPIC GAMES By Stavroula Bourna MASTER Thesis submitted to the professorial body for the partial fulfillment of obligations for the awarding of a post-graduate title in the Post-graduate Programme, "Organization and Management of Olympic Events" of the University of the Peloponnese, in the branch "Olympic Education" Sparta 2016 Approved by the Professor body: 1st Supervisor: Konstantinos Georgiadis Prof. UNIVERSITY OF PELOPONNESE, GREECE 2nd Supervisor: Konstantinos Mountakis Prof. UNIVERSITY OF PELOPONNESE, GREECE 3rd Supervisor: Paraskevi Lioumpi, Prof., GREECE ii Copyright © Stavroula Bourna, 2016 All rights reserved. The copying, storage and forwarding of the present work, either complete or in part, for commercial profit, is forbidden. The copying, storage and forwarding for non profit-making, educational or research purposes is allowed under the condition that the source of this information must be mentioned and the present stipulations be adhered to. Requests concerning the use of this work for profit-making purposes must be addressed to the author. The views and conclusions expressed in the present work are those of the writer and should not be interpreted as representing the official views of the Department of Sports’ Organization and Management of the University of the Peloponnese. iii ABSTRACT Stavroula Bourna: Technology Enhancement and Ethics in the Paralympic Games (Under the supervision of Konstantinos Georgiadis, Professor) The aim of the present thesis is to present how the new technological advances can affect the performance of the athletes in the Paralympic Games. -

Generalized CP Symmetries and Special Regions of Parameter Space in the Two-Higgs-Doublet Model
PHYSICAL REVIEW D 79, 116004 (2009) Generalized CP symmetries and special regions of parameter space in the two-Higgs-doublet model P.M. Ferreira,1,2 Howard E. Haber,3 and Joa˜o P. Silva1,4 1Instituto Superior de Engenharia de Lisboa, Rua Conselheiro Emı´dio Navarro, 1900 Lisboa, Portugal 2Centro de Fı´sica Teo´rica e Computacional, Faculdade de Cieˆncias, Universidade de Lisboa, Av. Prof. Gama Pinto 2, 1649-003 Lisboa, Portugal 3Santa Cruz Institute for Particle Physics, University of California, Santa Cruz, California 95064, USA 4Centro de Fı´sica Teo´rica de Partı´culas, Instituto Superior Te´cnico, P-1049-001 Lisboa, Portugal (Received 27 March 2009; published 15 June 2009) We consider the impact of imposing generalized CP symmetries on the Higgs sector of the two-Higgs- doublet model, and identify three classes of symmetries. Two of these classes constrain the scalar potential parameters to an exceptional region of parameter space, which respects either a Z2 discrete flavor symmetry or a Uð1Þ symmetry. We exhibit a basis-invariant quantity that distinguishes between these two possible symmetries. We also show that the consequences of imposing these two classes of CP symmetry can be achieved by combining Higgs family symmetries, and that this is not possible for the usual CP symmetry. We comment on the vacuum structure and on renormalization in the presence of these symmetries. Finally, we demonstrate that the standard CP symmetry can be used to build all the models we identify, including those based on Higgs family symmetries. DOI: 10.1103/PhysRevD.79.116004 PACS numbers: 11.30.Er, 11.30.Ly, 12.60.Fr, 14.80.Cp metry. -

CP CHEMICAL INDUSTRY PALLETS Edition: 7
July 2017 CP CHEMICAL INDUSTRY PALLETS Edition: 7 Foreword The European chemical and polymer industry is using a large amount of wooden pallets for the distribution of goods. For environmental, quality and safety reasons there is a strong need to organise the use and re-use of these pallets. Within PlasticsEurope, a team of experts from various chemicals and plastics producing companies, together with pallet specialists, have developed in the early ‘90 a standard for wooden pallets and drafted the present manufacturing and reconditioning specifications of the Chemical industry Pallets, called CP pallets. Special attention has been paid to quality, safety and environmental aspects. Although CP pallets have been designed for specific packages commonly used within the chemical and polymer industry, they are also suitable for other loads. Tests performed by various companies and testing institutes and experience from the use and re-use of millions of CP’s since 1991 have demonstrated that they conform to the needs of the chemical and polymer industry and their customers. However, PlasticsEurope Services cannot be held responsible for any problems or liabilities which may result from the use of the CP pallets system. Scope This document describes the pallet supplier registration and the system to collect used CP pallets. It defines the CP manufacturing, reconditioning and quality assurance criteria. It informs about the safe working load of CP’s. Correspondence can be addressed to: Avenue E. Van Nieuwenhuyse 4 Box 3 at 1160 Brussels, Belgium [email protected] www.plasticseurope.org Note: The content of this document is not essentially different from the previous edition, but has been completed with clarifications about the Supplier registration (header A.) and Marking (header B. -

Reconstruction from the Deck of K-Vertex Induced Subgraphs
Reconstruction from the deck of k-vertex induced subgraphs Hannah Spinoza∗ and Douglas B. West† Revised July 2018 Abstract The k-deck of a graph is its multiset of subgraphs induced by k vertices; we study what can be deduced about a graph from its k-deck. We strengthen a result of Manvel 2 by proving for ℓ ∈ N that when n is large enough (n > 2ℓ(ℓ+1) suffices), the (n − ℓ)- deck determines whether an n-vertex graph is connected (n ≥ 25 suffices when ℓ = 3, and n ≤ 2l cannot suffice). The reconstructibility ρ(G) of a graph G with n vertices is the largest ℓ such that G is determined by its (n − ℓ)-deck. We generalize a result of Bollob´as by showing ρ(G) ≥ (1 − o(1))n/2 for almost all graphs. As an upper bound on min ρ(G), we have ρ(Cn) = ⌈n/2⌉ = ρ(Pn) + 1. More generally, we compute ρ(G) whenever ∆(G) = 2, which involves extending a result of Stanley. Finally, we show that a complete r-partite graph is reconstructible from its (r + 1)-deck. MSC Codes: 05C60, 05C07 Key words: graph reconstruction, deck, reconstructibility, connected, random graph 1 Introduction A card of a graph G is a subgraph of G obtained by deleting one vertex. Cards are unlabeled, so only the isomorphism class of a card is given. The deck of G is the multiset of all cards of G. A graph is reconstructible if it is uniquely determined by its deck. The famous Reconstruction Conjecture was first posed in 1942. -

Publication 938 (Rev. November 2019) Introduction
Userid: CPM Schema: tipx Leadpct: 100% Pt. size: 8 Draft Ok to Print AH XSL/XML Fileid: … ons/P938/201911/A/XML/Cycle02/source (Init. & Date) _______ Page 1 of 120 10:35 - 19-Nov-2019 The type and rule above prints on all proofs including departmental reproduction proofs. MUST be removed before printing. Publication 938 (Rev. November 2019) Introduction Cat. No. 10647L Section references are to the Internal Revenue Department Code unless otherwise noted. of the This publication contains directories relating Treasury to real estate mortgage investment conduits Real Estate (REMICs) and collateralized debt obligations Internal (CDOs). The directory for each calendar quarter Revenue is based on information submitted to the IRS Service Mortgage during that quarter. For each quarter, there is a directory of new REMICs and CDOs and, if required, a section Investment containing amended listings. You can use the directory to find the representative of the RE- MIC or the issuer of the CDO from whom you Conduits can request tax information. The amended list- ing section shows changes to previously listed REMICs and CDOs. The update for each calen- (REMICs) dar quarter will be added to this publication ap- proximately six weeks after the end of the quar- Reporting ter. Publication 938 is only available on the In- ternet. To get Publication 938, including prior is- Information sues, visit IRS.gov. Future developments. The IRS has created a page on IRS.gov that includes information (And Other about Publication 938 at IRS.gov/Pub938. Infor- mation about any future developments affecting Collateralized Debt Publication 938 (such as legislation enacted af- Obligations (CDOs)) ter we release it) will be posted on that page. -

Publication 938 (Rev. December 1998) Introduction Cat
Publication 938 (Rev. December 1998) Introduction Cat. No. 10647L This publication contains directories relating Department to real estate mortgage investment conduits of the (REMICs) and collateralized debt obligations Treasury (CDOs), including CDOs issued in the form of “regular interests” in Financial Asset Internal Real Estate Securitization Investment Trusts (FASITs). Revenue The directory for each calendar quarter is Service based on information submitted to the Internal Mortgage Revenue Service during that quarter. This publication is only available on the IRS elec- tronic bulletin board and the Internet. Investment For each quarter, there is: • A directory of new REMICs and CDOs, and Conduits • A section containing amended listings. You can use the directory to find the repre- (REMICs) sentative of the REMIC or the issuer of the CDO from whom you can request tax infor- mation. The amended listing section shows changes to previously listed REMICs and Reporting CDOs. The directory for each calendar quarter will be added to this publication approximately Information six weeks after the end of the quarter. (And Other Other information. Publication 550, Invest- ment Income and Expenses, discusses the Collateralized Debt tax treatment that applies to holders of these investment products. For other information Obligations (CDOs)) about REMICs, see sections 860A through 860G of the Internal Revenue Code and any regulations issued under those sections. For information on FASITs, see Code sections 860H through 860L. Back issues. To get information for a REMIC or CDO with a startup/issue date prior to 1998 follow these procedures. Prior to 1996. Paper editions of Publica- tion 938 are available for periods prior to 1996. -

Dimension 4: Getting Something from Nothing
DIMENSION 4: GETTING SOMETHING FROM NOTHING RON STERN UNIVERSITY OF CALIFORNIA, IRVINE MAY 6, 2010 JOINT WORK WITH RON FINTUSHEL Topological n-manifold: locally homeomorphic to Rn TOPOLOGICAL VS. SMOOTH MANIFOLDS Topological n-manifold: locally homeomorphic to Rn VS Smooth n-manifold: locally diffeomorphic to Rn TOPOLOGICAL VS. SMOOTH MANIFOLDS Topological n-manifold: locally homeomorphic to Rn VS Smooth n-manifold: locally diffeomorphic to Rn Low dimensions: n=1,2,3 topological smooth ⇐⇒ TOPOLOGICAL VS. SMOOTH MANIFOLDS Topological n-manifold: locally homeomorphic to Rn VS Smooth n-manifold: locally diffeomorphic to Rn Low dimensions: n=1,2,3 topological smooth ⇐⇒ High dimensions: n > 4: smooth structures on topological mflds determined up to finite indeterminacy by characteristic classes of the tangent bundle TOPOLOGICAL VS. SMOOTH MANIFOLDS DIMENSION 4 Techniques from other dimensions do not apply (in fact fail in a rather dramatic fashion) DIMENSION 4 Techniques from other dimensions do not apply (in fact fail in a rather dramatic fashion) IN FACT IT COULD BE THE CASE THAT EVERY CLOSED TOPOLOGICAL MANIFOLD HAS EITHER NO OR INFINITELY MANY SMOOTH STRUCTURES! NO KNOWN COUNTEREXAMPLE DIMENSION 4 Techniques from other dimensions do not apply (in fact fail in a rather dramatic fashion) IN FACT IT COULD BE THE CASE THAT EVERY CLOSED TOPOLOGICAL MANIFOLD HAS EITHER NO OR INFINITELY MANY SMOOTH STRUCTURES! NO KNOWN COUNTEREXAMPLE HOW TO PROVE (OR DISPROVE) SUCH A WILD STATEMENT? FOR SIMPLICITY LET’S ASSUME SIMPLY-CONNECTED Classification of