Carbon Stable Isotope Composition of DNA Isolated from an Incipient Paleosol
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Ecological Site F136XY031GA PROVISIONAL - Bottomland Forest, Thermic Temperature Regime
Natural Resources Conservation Service Ecological site F136XY031GA PROVISIONAL - Bottomland Forest, Thermic Temperature Regime Accessed: 10/02/2021 General information MLRA notes Major Land Resource Area (MLRA): 136X–Southern Piedmont This area is in North Carolina (29 percent), Georgia (27 percent), Virginia (21 percent), South Carolina (16 percent), and Alabama (7 percent). It makes up about 64,395 square miles (166,865 square kilometers). (Ag Bulletin 296) The northeast-southwest trending Piedmont ecoregion comprises a transitional area between the mostly mountainous ecoregions of the Appalachians to the northwest and the relatively flat coastal plain to the southeast. It is a complex mosaic of Precambrian and Paleozoic metamorphic and igneous rocks with moderately dissected irregular plains and some hills. (EPA Ecoregions descriptions) ADD APPROPRIATE ECOREGION DESCRIPTION(S) Classification relationships A PROVISIONAL ECOLOGICAL SITE is a conceptual grouping of soil map unit components within a Major Land Resource Area (MLRA) based on the similarities in response to management. Although there may be wide variability in the productivity of the soils grouped into a Provisional Site, the soil vegetation interactions as expressed in the State and Transition Model are similar and the management actions required to achieve objectives, whether maintaining the existing ecological state or managing for an alternative state, are similar. Provisional Sites are likely to be refined into more precise group during the process of meeting the APPROVED ECOLOGICAL SITE DESCRIPTION criteria. This PROVISIONAL ECOLOGICAL SITE has been developed to meet the standards established in the National Ecological Site Handbook. The information associated with this ecological site does not meet the Approved Ecological Site Description Standard, but it has been through a Quality Control and Quality Assurance processes to assure consistency and completeness. -

Patterns of Flammability Across the Vascular Plant Phylogeny, with Special Emphasis on the Genus Dracophyllum
Lincoln University Digital Thesis Copyright Statement The digital copy of this thesis is protected by the Copyright Act 1994 (New Zealand). This thesis may be consulted by you, provided you comply with the provisions of the Act and the following conditions of use: you will use the copy only for the purposes of research or private study you will recognise the author's right to be identified as the author of the thesis and due acknowledgement will be made to the author where appropriate you will obtain the author's permission before publishing any material from the thesis. Patterns of flammability across the vascular plant phylogeny, with special emphasis on the genus Dracophyllum A thesis submitted in partial fulfilment of the requirements for the Degree of Doctor of philosophy at Lincoln University by Xinglei Cui Lincoln University 2020 Abstract of a thesis submitted in partial fulfilment of the requirements for the Degree of Doctor of philosophy. Abstract Patterns of flammability across the vascular plant phylogeny, with special emphasis on the genus Dracophyllum by Xinglei Cui Fire has been part of the environment for the entire history of terrestrial plants and is a common disturbance agent in many ecosystems across the world. Fire has a significant role in influencing the structure, pattern and function of many ecosystems. Plant flammability, which is the ability of a plant to burn and sustain a flame, is an important driver of fire in terrestrial ecosystems and thus has a fundamental role in ecosystem dynamics and species evolution. However, the factors that have influenced the evolution of flammability remain unclear. -

NLI Recommended Plant List for the Mountains
NLI Recommended Plant List for the Mountains Notable Features Requirement Exposure Native Hardiness USDA Max. Mature Height Max. Mature Width Very Wet Very Dry Drained Moist &Well Occasionally Dry Botanical Name Common Name Recommended Cultivars Zones Tree Deciduous Large (Height: 40'+) Acer rubrum red maple 'October Glory'/ 'Red Sunset' fall color Shade/sun x 2-9 75' 45' x x x fast growing, mulit-stemmed, papery peeling Betula nigra river birch 'Heritage® 'Cully'/ 'Dura Heat'/ 'Summer Cascade' bark, play props Shade/part sun x 4-8 70' 60' x x x Celtis occidentalis hackberry tough, drought tolerant, graceful form Full sun x 2-9 60' 60' x x x Fagus grandifolia american beech smooth textured bark, play props Shade/part sun x 3-8 75' 60' x x Fraxinus americana white ash fall color Full sun/part shade x 3-9 80' 60' x x x Ginkgo biloba ginkgo; maidenhair tree 'Autumn Gold'/ 'The President' yellow fall color Full sun 3-9 70' 40' x x good dappled shade, fall color, quick growing, Gleditsia triacanthos var. inermis thornless honey locust Shademaster®/ Skyline® salt tolerant, tolerant of acid, alkaline, wind. Full sun/part shade x 3-8 75' 50' x x Liriodendron tulipifera tulip poplar fall color, quick growth rate, play props, Full sun x 4-9 90' 50' x Platanus x acerifolia sycamore, planetree 'Bloodgood' play props, peeling bark Full sun x 4-9 90' 70' x x x Quercus palustris pin oak play props, good fall color, wet tolerant Full sun x 4-8 80' 50' x x x Tilia cordata Little leaf Linden, Basswood 'Greenspire' Full sun/part shade 3-7 60' 40' x x Ulmus -

Native Tree Families, Including Large and Small Trees, 1/1/08 in the Southern Blue Ridge Region (Compiled by Rob Messick Using Three Sources Listed Below.)
Native Tree Families, Including Large and Small Trees, 1/1/08 in the Southern Blue Ridge Region (Compiled by Rob Messick using three sources listed below.) • Total number of tree families listed in the southern Blue Ridge region = 33. • Total number of native large and small tree species listed = 113. (Only 84 according to J. B. & D. L..) There are 94 tree species in more frequently encountered families. There are 19 tree species in less frequently encountered families. • There is 93 % compatibility between Ashe & Ayers (1902), Little (1980), and Swanson (1994). (W. W. Ashe lists 105 tree species in the region in 1902. These are fully compatible with current listings.) ▸means more frequently encountered species. ?? = means a tree species that possibly occurs in the region, though its presence is not clear. More frequently encountered tree families (21): Pine Family Cashew Family Walnut Family Holly Family Birch Family Maple Family Beech Family Horse-chestnut (Buckeye) Family Magnolia Family Linden (Basswood) Family Laurel Family Tupelo-gum Family Witch-hazel Family Dogwood Family Plane-tree (Sycamore) Family Heath Family Rose Family Ebony Family Legume Family Storax (Snowbell) Family Olive Family Less frequently encountered tree families (12): Cypress Family Bladdernut Family Willow Family Buckthorn Family Elm Family Tea Family Mulberry Family Ginseng Family Custard-apple (Annona) Family Sweetleaf Family Rue Family Honeysuckle Family ______________________________________________________________________________ • More Frequently Encountered Tree Families: Pine Family (10): ▸ Fraser fir - Abies fraseri (a.k.a. Balsam) ▸ red spruce - Picea rubens ▸ shortleaf pine - Pinus echinata ▸ table mountain pine - Pinus pungens ▸ pitch pine - Pinus rigida ▸ white pine - Pinus strobus ▸ Virginia pine - Pinus virginiana loblolly pine - Pinus taeda ▸ eastern hemlock - Tsuga canadensis (a.k.a. -

Camellia Sinensis (L.) Kuntze (7)
“As Primeiras Camélias Asiáticas a Chegarem a Portugal e à Europa”. Armando Oliveira António Sanches (1623), Planisfério. 1 O género Camellia L. está praticamente confinado ao sul da China (80% de todas as espécies) e à região do sul da Ásia que inclui as Filipinas e as zonas do noroeste do arquipélago da Indonésia, com a inclusão do Japão e partes da Coreia. Estima-se que praticamente 20% das espécies de Camellia se encontram no Vietname. A região fitogeográfica do sul da Ásia é composta pela China, Laos, Mianmar (ex-Birmânia), Tailândia, Camboja e Vietname. 1 (Huang et al., 2016) 106 • A proposta taxonómica de Linnaeus (1835), “Sistema Natura”, permitiu-nos obter uma mais fácil e rápida identificação das espécies. • Baseia-se numa classificação dita binomial que atribui nomes compostos por duas palavras, quase sempre recorrendo ao latim. Adaptado de Fairy Lake Botanical Garden Flora (2018) 2 Reino Filo Classe Ordem Família Género Espécies/Variedades Cultivares Camellia caudata Wall. (11) Camellia drupifera Lour. (4) Dicotiledóneas Antófitas Camellia euryoides Lindl. (7) Vegetal (a semente Ericales (25) Theaceaes (12) Camellia (102+40) (que dão flor) contém 2 ou mais Camellia japonica L. cotilédones) Camellia kissi Wall. (11) Camellia oleifera Abel (6) Camellia rosaeflora Hook. (1) Camellia sasanqua Thunb. Camellia sinensis (L.) Kuntze (7) • A 1ª parte do nome é referente ao género da espécie em causa e a 2ª parte identifica a espécie dentro de um determinado género. Adaptado de Fairy Lake Botanical Garden Flora (2018) 2 Ordem Família -

Ornamental Shrubs and Groundcovers
ORNAMENTAL SHRUBS AND GROUNDCOVERS University of California Cooperative Extension Farm and Home Advisors 1031 S. Mt. Vernon Avenue ● Bakersfield, CA 93307 This list is intended to be a guide. Plant availability may change and improved varieties may come to market. Quantitative data do not exist for plant water use for most species. Sensitivity to possible plant allergens varies among people. AUTHOR John Karlik, Advisor, Environmental Horticulture/Environmental Science and the following: Peter Brown, Earth Landscape, Ridgecrest, CA John Karnes, Klassen Corporation Robert Martin, North of the River Recreation and Park District Mitchell Perez, Kern High School District Steph Sanders, North of the River Recreation and Park District Alfonso Valadez, Kern High School District Revised and expanded, August 2020 REFERENCES Duffield, Mary R. and Jones, Warren D. 1981. Plants for Dry Climates: How to Select, Grow and Enjoy. H.P. Publishing, Tucson, AZ Perry, Bob. 1981. Trees and Shrubs for Dry California Landscapes. Land Design Publishing, San Dimas, CA Sunset New Western Garden Book The University of California, Division of Agriculture and Natural Resources (UC ANR) prohibits discrimination against or harassment of any person employed by or seeking employment with UC ANR on the basis of race, color, national origin, religion, sex, gender, gender expression, gender identity, pregnancy (which includes pregnancy, childbirth, and medical conditions related to pregnancy or childbirth), physical or mental disability, medical condition (cancer-related or genetic characteristics), genetic information (including family medical history), ancestry, marital status, age, sexual orientation, citizenship, status as a protected veteran or service in the uniformed services (as defined by the Uniformed Services Employment and Reemployment Rights Act of 1994 [USERRA]), as well as state military and naval service. -

Camellia Queen of the Southern Garden
PLANT GUIDES CAMELLIA Queen of the Southern Garden hen Harry P. Leu gave his gardens to and newly discovered species. Thousands of camellias the City of Orlando in 1961, it was said are what make Leu Gardens unique and the array of that his Camellia assortment contained shapes, colors and sizes provide outstanding color from more than 1,500 individual plants. December through March. Guests are encouraged to WToday the internationally recognized camellia collection visit during this time to enjoy the beautiful Central continues to expand and include many historic varieties Florida weather and the Leu’s garden. Camellia sasanqua: ‘Alabama Beauty’ ‘Cotton Candy’ ‘Daydream’ ‘Fuji-no-mine’ ‘Mine-no-yuki’ Deep Pink Pink Pink and White White White ‘Pink Dauphin’ ‘Setsugekka’ ‘Cleopatra’ ‘Maiden’s Blush’ ‘Stephanie Golden’ Pink White Pink Blush Pink Hot Pink Camellias — a Primer here are tens of thousands of registered Camellias at Leu Gardens cultivars of camellias and there are Camellia sasanqua Camellia japonica more than 200 different species. n Small, often simple flowers n Very large flowers TThey are all originally native to eastern and southeastern Asia with most native to China. n Wider, open type of growth n Upright growth habit The majority of Leu Gardens’ collection n Full sun to partial shade n Prefers partial to full shade consists of cultivars of Camellia japonica and n Bloom October through n Bloom late December Camellia sasanqua. Sasanqua camellias bloom December through March earlier than the C. japonica types (often from n Nearly all fragrant n Most varieties not fragrant October through December) and most prefer full sun. -
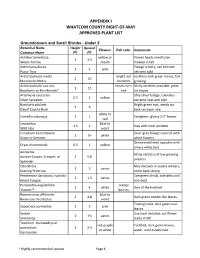
Whatcom County's Approved Plant List
APPENDIX I WHATCOM COUNTY RIGHT-OF-WAY APPROVED PLANT LIST Groundcovers and Small Shrubs - Under 2' Botanical Name Height Spread Flowers Fall color Comments Common Name (ft) (ft) Achillea tomentosa yellow or Flower heads need to be 1 3-5 Wooly Yarrow cream mowed in fall Antennaria dioica Foliage is furry, can tolerate 1 2 pink Pussy Toes extreme cold Arctostaphylos media bright red Leathery dark green leaves, fast 2 10 Manzanita Media branches growing Arctostaphylos uva ursi leaves turn Many varieties available, great 1 15 Bearberry or Kinnikinnick* red for slopes Artemesia caucasica Silky silver foliage, tolerates 0.5 2 yellow Silver Spreader extreme heat and cold Baccharis pilularis Bright green mat, needs cut 2 6 Dwarf Coyote Bush back once per year white to Camellia sasanqua 1 1 Evergreen, glossy 2-3" leaves red Ceanothus blue to 1.5 2 Stay with local varieties Wild Lilac violet Cerastium tomentosum Silver gray foliage covered with 1 5+ white Snow-in-Summer white flowers Ornamental seed capsules with Dryas drummondii 0.5 1 yellow silvery white tails Juniperus Many varieties of low growing Juniper-Carpet, Creeper, or 2 5-8 junipers Spreader Oenothera May die back in severe winters, 1 2 varies Evening Primrose come back strong Penstemon davidsonii, rupicola Evergreen shrub, tolerates cold 2 1.5 varies Beard Tongue and heat Pyracantha augustifolia orange 1 4 white One of the hardiest 'Gnome'* berries Rosmariunus officinalis blue to 2 4-8 Dark green needle-like leaves Rosemary 'Prostratus' violet Trailing habit, dark green oval Saponaria ocymoides 1 3 pink leaves Sedum Use local varieties, cut flower 2 1-5 varies Stonecrop stalks in fall Teucrium chamaedrys or red-purple Toothed, dark green leaves, postratium 1 2-3 or white water until established Germander * Highly-recommended species Page 1 Thymus white to 2 1-2 Many varieties Thyme purple Shrubs Botanical Name Height Spread Flowers Fall color Comments Common Name (ft) (ft) Arctostaphylos bright red Shiny dark green leaves turn 2-15 20 varies Manzanita berries maroon in fall. -

Uncultivated Nuts of the United States
Uncultivated Nuts of Forest Service The United States Agriculture Information Bulletin Number 450 Arnold Krochmal Connie Krochmal Pî f3 3 i-O CO D OJ U TJ 1 ■> li> en 0 h- < 0 CM c 0 o £ Q -ce ■y^ CCÎ CD .si <r CO T— ■¿i a _,i 0 CO < 0 0» D 2: I— HJ Figure 1.—Cherokee woman pounding nuts with wooden mortar and pestle (courtesy of the Smithsonian Institution, Neg. No. 55, 445). Uncultivated Nuts of The United States by Arnold Krochmal, Economic Botanist, Southeastern Forest Experiment Station, Asheville, N.C., and Connie Krochmal, Volunteer Writer United States Department of Agriculture Forest Service june 1982 Library of Congress Catalog Card Number: 81-600067 Krochmal, Arnold; Krochmal, Connie. Uncultivated nuts of the United States. Agrie. Info. Bull. 450. Washington, DC : U.S. Department of Agri- culture; 1982.89 p. A handbook of nuts and nutlike seeds from trees and shrubs that are not cultivated in orchards but generally grow wild or that are sometimes grown as ornamentals. Descriptions, illustrations, and range maps help the reader to identify species. The nuts and seeds are also discussed as a renewable resource of present or potential use to human beings. KEYWORDS: Nutrition, renewable resources. Acknowledgments Study materials and samples were provided by David Stoller of the Cornell Plantations and Harold Grelen, Lisle Green, Franklin Bonner, Rudolph Strothmann, Douglas Roy, and David Priester, all of the Forest Service. Herbarium specimens and information on distribution and nomenclature were made available by Frederick Meyer and Peter Mazzeo, U.S. National Arboretum; Stanwyn Shetler, Smithsonian Institution; Carol Keener, Pennsylvania State University; Gene Wofford, University of Tennessee; Daniel Ward, University of Florida; John Freeman, Auburn University; L. -

Cape Cod & Traditional
CAPE COD & TRADITIONAL 4/3/2019 Note: The following plants may not be used: Eucalyptus, London Plane Tree, Purple Leaf Plum Water BOTANICAL NAME COMMON NAME Height Evergreen Use Trees Alnus rhombifolia White Alder 90' No High Bauhinia blakeana 20' Yes Medium Betula pendula European White Birch 40' No Medium Cinnamomum camphora Camphor Tree 50' Yes Low Cupaniopsis anacardioides Carrotwood 40' Yes High Fraxinus velutina Modesto Ash 50' No Medium Persea americana Avodado 30' Yes Low Geijera parviflora Australian Willow 30' Yes Medium Gleditsia triacanthos 'inermis' 40' No Medium Jacaranda mimosiflora Jacaranda 40' No Medium Koelreuteria bipinnata Chinese Flame Tree 35' No Medium Koelreuteria panniculata Golden Rain Tree 30' No Medium Lagerstroemia indica Crape Myrtle 30' Yes Medium Laurus nobilis Sweet Bay 40' Yes Low Liquidambar styraciflua Sweet Gum 60' No Medium Magnolia grandiflora Southern Magnolia 80' Yes Medium Magnolia 'Samual Sommer' 40' Yes Medium Pinus canariensis Canary Island Pine 70' Yes Low Pinus aldarica Afghan Pine 80' Yes Medium Pinus halpensis Aleppo Pine 60' Yes Medium Pinus pinea Italian Stone Pine 80' Yes Medium Pistacia chinensis Chinese Pistachio 60' No Medium Platanus racemosa California Sycamore 100' No Medium Prunus s. 'Kwanzan' Japanese Flowering Cheerry 30' No Medium Podocarpus gracilior Fern Pine 60' Yes Medium Pyrus calleriana Ornamental Pear 40' No Medium Quercus ilex Holly Oak 60' Yes Low Quercus suber Cork Oak 70' Yes Low Quercus virginiana Souther Live Oak 60' Yes Medium Sophora japonica Japanese Pagoda -

Vegetation Community Monitoring at Congaree National Park: 2014 Data Summary
National Park Service U.S. Department of the Interior Natural Resource Stewardship and Science Vegetation Community Monitoring at Congaree National Park 2014 Data Summary Natural Resource Data Series NPS/SECN/NRDS—2016/1016 ON THIS PAGE Tiny, bright yellow blossoms of Hypoxis hirsuta grace the forest floor at Congaree National Park. Photograph courtesy of Sarah C. Heath, Southeast Coast Network. ON THE COVER Spiraling compound leaf of green dragon (Arisaema dracontium) at Congaree National Park. Photograph courtesy of Sarah C. Heath, Southeast Coast Network Vegetation Community Monitoring at Congaree National Park 2014 Data Summary Natural Resource Data Series NPS/SECN/NRDS—2016/1016 Sarah Corbett Heath1 and Michael W. Byrne2 1National Park Service Southeast Coast Inventory and Monitoring Network Cumberland Island National Seashore 101 Wheeler Street Saint Marys, GA 31558 2National Park Service Southeast Coast Inventory and Monitoring Network 135 Phoenix Drive Athens, GA 30605 May 2016 U.S. Department of the Interior National Park Service Natural Resource Stewardship and Science Fort Collins, Colorado The National Park Service, Natural Resource Stewardship and Science office in Fort Collins, Colorado, publishes a range of reports that address natural resource topics. These reports are of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Data Series is intended for the timely release of basic data sets and data summaries. Care has been taken to assure accuracy of raw data values, but a thorough analysis and interpretation of the data has not been completed. -

JBG Plant List
Jensen Botanical Garden Plant List When you are at the garden, you will see metal signs with labels. each will have a number and the common name of the plant. This list will give you more information. 1.Valley Oak; Quercus lobata. Native to the interior valleys and sierra foothills. Deciduous. California's mightiest oak, often reaching 70 feet or more, with equal or greater spread. This was on the property when Mr. Jensen bought it and has been estimated to be over 400 years old. 2. Rose; “Day Dream” Shrub. 18-24 inches high and 24-36 inches wide is normal. JBG ones are well established and very happy, so they exceed the normal size. Medium pink flower with a slight fragrance, multiple flowers on each stem. Blooms repeatedly - prune frequently to encourage new blooms. 3. Japanese Maple, Acer palatum “Sangu Kaku” or Senkaki. Vigorous, upright treelike growth. Fall foliage yellow, tinted rose. Twigs, branches striking coral red. 4. Flowering Maple, Abutilon. Evergreen vine shrubs. Part shade. Grown daily for bell shaped flowers. Bloom April-June, but some bloom year round. Available in white, yellow, pink, and red. Attracts bees and hummingbirds. 5. Sage, Salvia microphylla. Blooms all through the hottest summers, attracting hummingbirds and butterflies. Drought tolerant once established. Plant in full sun, grows quickly to 3 foot by 3 foot. 6. Interior live oak; Quercus wislizenii. Native to the Sierra foothills and east side of California’s central valley. Evergreen. Grows 30 - 75 feet high; often broader than high. Oblong, glossy green leaves to 4 inches long, smooth or spiny edged.