The Emerging Role of Lysine Methyltransferase SETD8 in Human
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Novel TAL1 Targets Beyond Protein-Coding Genes: Identification of TAL1-Regulated Micrornas in T-Cell Acute Lymphoblastic Leukemia
Letters to the Editor 1603 REFERENCES 8 Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R et al. Frequent 1 Rozman C, Montserrat E. Chronic lymphocytic leukemia. N Engl J Med 1995; 333: pathway mutations of splicing machinery in myelodysplasia. Nature 2011; 478: 64–69. 1052–1057. 9 Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D et al. 2 Zenz T, Mertens D, Kuppers R, Dohner H, Stilgenbauer S. From pathogenesis to Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med treatment of chronic lymphocytic leukaemia. Nat Rev Cancer 2010; 10: 37–50. 2011; 365: 1384–1395. 3 Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N et al. Whole- 10 Damm F, Nguyen-Khac F, Fontenay M, Bernard OA. Spliceosome and other novel genome sequencing identifies recurrent mutations in chronic lymphocytic leu- mutations in chronic lymphocytic leukemia and myeloid malignancies. Leukemia kaemia. Nature 2011; 475: 101–105. 2012; 26: 2027–2031. 4 Quesada V, Conde L, Villamor N, Ordonez GR, Jares P, Bassaganyas L et al. Exome 11 Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in RNP machine. Cell 2009; 136: 701–718. chronic lymphocytic leukemia. Nat Genet 2012; 44: 47–52. 12 David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways 5 Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K et al. SF3B1 and programs unhinged. Genes Dev 2010; 24: 2343–2364. -

University of California, San Diego
UNIVERSITY OF CALIFORNIA, SAN DIEGO The post-terminal differentiation fate of RNAs revealed by next-generation sequencing A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Biomedical Sciences by Gloria Kuo Lefkowitz Committee in Charge: Professor Benjamin D. Yu, Chair Professor Richard Gallo Professor Bruce A. Hamilton Professor Miles F. Wilkinson Professor Eugene Yeo 2012 Copyright Gloria Kuo Lefkowitz, 2012 All rights reserved. The Dissertation of Gloria Kuo Lefkowitz is approved, and it is acceptable in quality and form for publication on microfilm and electronically: __________________________________________________________________ __________________________________________________________________ __________________________________________________________________ __________________________________________________________________ __________________________________________________________________ Chair University of California, San Diego 2012 iii DEDICATION Ma and Ba, for your early indulgence and support. Matt and James, for choosing more practical callings. Roy, my love, for patiently sharing the ups and downs of this journey. iv EPIGRAPH It is foolish to tear one's hair in grief, as though sorrow would be made less by baldness. ~Cicero v TABLE OF CONTENTS Signature Page .............................................................................................................. iii Dedication .................................................................................................................... -

DNA·RNA Triple Helix Formation Can Function As a Cis-Acting Regulatory
DNA·RNA triple helix formation can function as a cis-acting regulatory mechanism at the human β-globin locus Zhuo Zhoua, Keith E. Gilesa,b,c, and Gary Felsenfelda,1 aLaboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892; bUniversity of Alabama at Birmingham Stem Cell Institute, University of Alabama at Birmingham, Birmingham, AL 35294; and cDepartment of Biochemistry and Molecular Genetics, University of Alabama at Birmingham, Birmingham, AL 35294 Contributed by Gary Felsenfeld, February 4, 2019 (sent for review January 4, 2019; reviewed by James Douglas Engel and Sergei M. Mirkin) We have identified regulatory mechanisms in which an RNA tran- of the criteria necessary to establish the presence of a triplex script forms a DNA duplex·RNA triple helix with a gene or one of its structure, we first describe and characterize triplex formation at regulatory elements, suggesting potential auto-regulatory mecha- the FAU gene in human erythroid K562 cells. FAU encodes a nisms in vivo. We describe an interaction at the human β-globin protein that is a fusion containing fubi, a ubiquitin-like protein, locus, in which an RNA segment embedded in the second intron of and ribosomal protein S30. Although fubi function is unknown, the β-globin gene forms a DNA·RNA triplex with the HS2 sequence posttranslational processing produces S30, a component of the within the β-globin locus control region, a major regulator of glo- 40S ribosome. We used this system to refine methods necessary bin expression. We show in human K562 cells that the triplex is to detect triplex formation and to distinguish it from R-loop stable in vivo. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Transcritional Regulatory Networks Downstream of TAL1/SCL in T-Cell Acute Lymphoblastic Leukemia
Supplemental Materials Transcritional Regulatory Networks Downstream of TAL1/SCL in T-cell Acute Lymphoblastic Leukemia Palomero et al. Figure S1. Downregulation of TAL1 by RNA interference does not affect apoptosis in Jurkat cells. Figure S2. Scatter plots of Chromatin immunoprecipitations performed with TAL1#370 antibody in Jurkat cells. Figure S3. Target validation by gene-specific quantitative RT-PCR (qRT-PCR) Table S1. Genes regulated by TAL1 knockdown are identified as direct targets of this transcription factor using ChIP on chip. Table S2. Analysis of TAL1, E2A and HEB binding to TAL1 direct targets identified by ChIP on chip. Table S3. Previously described TAL1 targets. Figure S1. Downregulation of TAL1 by RNA interference does not affect apoptosis in Jurkat cells. Annexin V staining was used to quantify apoptosis rates in Jurkat cell clones expressing TAL1 shRNA (right panels) or a control shRNA (left panels). The percentage of apoptotic cells is indicated in the bottom right corner of each graph, while the dead cell percentage is indicated in the top right corner. PI: propidium iodide. Name Description p-value IFRD1 interferon-related developmental regulator 1 0.009927703 PCK2 phosphoenolpyruvate carboxykinase 2 (mitochondrial) 0.00445631 ATRX alpha thalassemia/mental retardation syndrome X-linked (RAD54 homolog, S. cerevisiae) 0.010847575 CDK6 cyclin-dependent kinase 6 0.044093956 Table S1. Genes regulated by TAL1 knockdown are identified as direct target of this transcription factor using ChIP on chip. The p-value determined by the error model applied to the ChIP on chip fluorescence data is indicated in the right column. Name Description Name Description E2A E2A HEB HEB TAL1 TAL1 Signal transduction-Receptor Transporters-lipids/small molecules MUC16 mucin 16 ABCC12 ATP-binding cassette, sub-fam. -

Function of Bromodomain and Extra-Terminal Motif Proteins (Bets) in Gata1-Mediated Transcription
University of Pennsylvania ScholarlyCommons Publicly Accessible Penn Dissertations 2015 Function of Bromodomain and Extra-Terminal Motif Proteins (bets) in Gata1-Mediated Transcription Aaron James Stonestrom University of Pennsylvania, [email protected] Follow this and additional works at: https://repository.upenn.edu/edissertations Part of the Molecular Biology Commons, and the Pharmacology Commons Recommended Citation Stonestrom, Aaron James, "Function of Bromodomain and Extra-Terminal Motif Proteins (bets) in Gata1-Mediated Transcription" (2015). Publicly Accessible Penn Dissertations. 1148. https://repository.upenn.edu/edissertations/1148 This paper is posted at ScholarlyCommons. https://repository.upenn.edu/edissertations/1148 For more information, please contact [email protected]. Function of Bromodomain and Extra-Terminal Motif Proteins (bets) in Gata1-Mediated Transcription Abstract Bromodomain and Extra-Terminal motif proteins (BETs) associate with acetylated histones and transcription factors. While pharmacologic inhibition of this ubiquitous protein family is an emerging therapeutic approach for neoplastic and inflammatory disease, the mechanisms through which BETs act remain largely uncharacterized. Here we explore the role of BETs in the physiologically relevant context of erythropoiesis driven by the transcription factor GATA1. First, we characterize functions of the BET family as a whole using a pharmacologic approach. We find that BETs are broadly required for GATA1-mediated transcriptional activation, but that repression is largely BET-independent. BETs support activation by facilitating both GATA1 occupancy and transcription downstream of its binding. Second, we test the specific olesr of BETs BRD2, BRD3, and BRD4 in GATA1-activated transcription. BRD2 and BRD4 are required for efficient anscriptionaltr activation by GATA1. Despite co-localizing with the great majority of GATA1 binding sites, we find that BRD3 is not equirr ed for GATA1-mediated transcriptional activation. -

DNA Methylation Changes in Down Syndrome Derived Neural Ipscs Uncover Co-Dysregulation of ZNF and HOX3 Families of Transcription
Laan et al. Clinical Epigenetics (2020) 12:9 https://doi.org/10.1186/s13148-019-0803-1 RESEARCH Open Access DNA methylation changes in Down syndrome derived neural iPSCs uncover co- dysregulation of ZNF and HOX3 families of transcription factors Loora Laan1†, Joakim Klar1†, Maria Sobol1, Jan Hoeber1, Mansoureh Shahsavani2, Malin Kele2, Ambrin Fatima1, Muhammad Zakaria1, Göran Annerén1, Anna Falk2, Jens Schuster1 and Niklas Dahl1* Abstract Background: Down syndrome (DS) is characterized by neurodevelopmental abnormalities caused by partial or complete trisomy of human chromosome 21 (T21). Analysis of Down syndrome brain specimens has shown global epigenetic and transcriptional changes but their interplay during early neurogenesis remains largely unknown. We differentiated induced pluripotent stem cells (iPSCs) established from two DS patients with complete T21 and matched euploid donors into two distinct neural stages corresponding to early- and mid-gestational ages. Results: Using the Illumina Infinium 450K array, we assessed the DNA methylation pattern of known CpG regions and promoters across the genome in trisomic neural iPSC derivatives, and we identified a total of 500 stably and differentially methylated CpGs that were annotated to CpG islands of 151 genes. The genes were enriched within the DNA binding category, uncovering 37 factors of importance for transcriptional regulation and chromatin structure. In particular, we observed regional epigenetic changes of the transcription factor genes ZNF69, ZNF700 and ZNF763 as well as the HOXA3, HOXB3 and HOXD3 genes. A similar clustering of differential methylation was found in the CpG islands of the HIST1 genes suggesting effects on chromatin remodeling. Conclusions: The study shows that early established differential methylation in neural iPSC derivatives with T21 are associated with a set of genes relevant for DS brain development, providing a novel framework for further studies on epigenetic changes and transcriptional dysregulation during T21 neurogenesis. -

Targeting the Methyltransferase SETD8 Impairs Tumor Cell Survival and Overcomes Drug Resistance Independently of P53 Status in Multiple Myeloma
bioRxiv preprint doi: https://doi.org/10.1101/776930; this version posted September 20, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Targeting the methyltransferase SETD8 impairs tumor cell survival and overcomes drug resistance independently of p53 status in multiple myeloma Laurie Herviou (1,2,4), Fanny Izard (3,4), Ouissem Karmous-Gadacha (2) , Claire Gourzones (1), Celine Bellanger (1), Eva Desmedt (5), Anqi Ma (6), Laure Vincent (7) , Guillaume Cartron (4,7), Karin Vanderkerken (5), Jian Jin (6), Elke De Bruyne (5), Charlotte Grimaud (3,4,8), Eric Julien (3,4,8 +) and Jérôme Moreaux (1,2,4+) (1) IGH, CNRS, Univ Montpellier, France (2) CHU Montpellier, Laboratory for Monitoring Innovative Therapies, Department of Biological Hematology, Montpellier, France (3) Institut de Recherche en Cancérologie de Montpellier (IRCM), INSERM U1194, Institut Régional du Cancer (ICM), Montpellier F-34298, France (4) University of Montpellier, Montpellier, F-34090, France (5) Department of Hematology and Immunology-Myeloma Center Brussels, Vrije Universiteit Brussel, Brussels, Belgium (6) Mount Sinai Center for Therapeutics Discovery, Departments of Pharmacological Sciences and Oncological Sciences, Tisch Cancer Institute, Icahn School of Medicine at Mount Sinai, NeW York, NeW York 10029, United States. (7) CHU Montpellier, Department of Clinical Hematology, Montpellier, France (8) Centre National de la Recherche Scientifique (CNRS), F-34293, Montpellier, France + : co-last and corresponding authors ; corresponding authors: Eric Julien ([email protected]) and jérôme Moreaux ([email protected]). 1 bioRxiv preprint doi: https://doi.org/10.1101/776930; this version posted September 20, 2019. -

TRCP Promotes Cell Growth by Targeting PR-Set7/Set8 for Degradation
SCFβ-TRCP promotes cell growth by targeting PR-Set7/Set8 for degradation The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Wang, Z., X. Dai, J. Zhong, H. Inuzuka, L. Wan, X. Li, L. Wang, et al. 2015. “SCFβ-TRCP promotes cell growth by targeting PR-Set7/Set8 for degradation.” Nature Communications 6 (1): 10185. doi:10.1038/ ncomms10185. http://dx.doi.org/10.1038/ncomms10185. Published Version doi:10.1038/ncomms10185 Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:23993465 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA ARTICLE Received 4 May 2015 | Accepted 12 Nov 2015 | Published 15 Dec 2015 DOI: 10.1038/ncomms10185 OPEN SCFb-TRCP promotes cell growth by targeting PR-Set7/Set8 for degradation Zhiwei Wang1,2,*, Xiangpeng Dai2,*, Jiateng Zhong2,3,*, Hiroyuki Inuzuka2, Lixin Wan2, Xiaoning Li2,4, Lixia Wang1, Xiantao Ye1, Liankun Sun4, Daming Gao2,5,LeeZou6 & Wenyi Wei2 The Set8/PR-Set7/KMT5a methyltransferase plays critical roles in governing transcriptional regulation, cell cycle progression and tumorigenesis. Although CRL4Cdt2 was reported to regulate Set8 stability, deleting the PIP motif only led to partial resistance to ultraviolet- induced degradation of Set8, indicating the existence of additional E3 ligase(s) controlling Set8 stability. Furthermore, it remains largely undefined how DNA damage-induced kinase cascades trigger the timely destruction of Set8 to govern tumorigenesis. -
H4K20 Monomethylation Faces the WNT
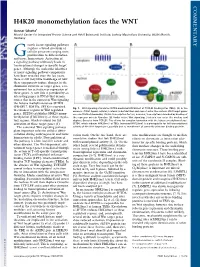
COMMENTARY H4K20 monomethylation faces the WNT Gunnar Schotta1 Munich Center for Integrated Protein Science and Adolf Butenandt Institute, Ludwig Maximilians University, 80336 Munich, Germany rowth factor signaling pathways regulate a broad spectrum of G cellular processes ranging from proliferation to differentiation and tissue homeostasis. Activation of a signaling pathway ultimately leads to transcriptional changes in specific target genes. Although the molecular identities of many signaling pathway components have been revealed over the last years, there is still very little knowledge of how these components induce changes in the chromatin structure of target genes, a re- quirement for activation or repression of these genes. A new link is provided by an interesting paper in PNAS that demon- strates that in the context of Wnt signaling the histone methyltransferase SETD8 (PR-SET7, KMT5a, SET8) is recruited Fig. 1. Wnt signaling stimulates SETD8-mediated H4K20me1 at TCF/LEF binding sites (TBEs). (A)Inthe to enhancer regions of Wnt-regulated absence of Wnt ligand, cellular β-catenin is destabilized and cannot enter the nucleus. Wnt target genes genes. SETD8 establishes H4K20 mono- are constitutively bound by TCF/LEF transcription factors; however, transcription is blocked by binding of methylation (H4K20me1) at these regula- the repressor protein Groucho. (B) Under active Wnt signaling, β-catenin can enter the nucleus and tory regions, which is crucial for full displace Groucho from TCF/LEF. This allows for complex formation with the histone methyltransferase activation of these target genes (1). SETD8, which induces H4K20me1 at TBEs. Increased H4K20me1 is a prerequisite for full transcriptional The canonical Wnt signaling pathway activity of the Wnt target gene, possibly due to recruitment of currently unknown binding proteins. -

Sleeping Beauty Transposon Mutagenesis Identifies Genes That
Sleeping Beauty transposon mutagenesis identifies PNAS PLUS genes that cooperate with mutant Smad4 in gastric cancer development Haruna Takedaa,b, Alistair G. Rustc,d, Jerrold M. Warda, Christopher Chin Kuan Yewa, Nancy A. Jenkinsa,e, and Neal G. Copelanda,e,1 aDivision of Genomics and Genetics, Institute of Molecular and Cell Biology, Agency for Science, Technology and Research, Singapore 138673; bDepartment of Pathology, School of Medicine, Kanazawa Medical University, Ishikawa 920-0293, Japan; cExperimental Cancer Genetics, Wellcome Trust Sanger Institute, Cambridge CB10 1HH, United Kingdom; dTumour Profiling Unit, The Institute of Cancer Research, Chester Beatty Laboratories, London SW3 6JB, United Kingdom; and eCancer Research Program, Houston Methodist Research Institute, Houston, TX 77030 Contributed by Neal G. Copeland, February 27, 2016 (sent for review October 15, 2015; reviewed by Yoshiaki Ito and David A. Largaespada) Mutations in SMAD4 predispose to the development of gastroin- animal models that mimic human GC, researchers have infected testinal cancer, which is the third leading cause of cancer-related mice with H. pylori and then, treated them with carcinogens. They deaths. To identify genes driving gastric cancer (GC) development, have also used genetic engineering to develop a variety of trans- we performed a Sleeping Beauty (SB) transposon mutagenesis genic and KO mouse models of GC (10). Smad4 KO mice are one + − screen in the stomach of Smad4 / mutant mice. This screen iden- GC model that has been of particular interest to us (11, 12). tified 59 candidate GC trunk drivers and a much larger number of Heterozygous Smad4 KO mice develop polyps in the pyloric re- candidate GC progression genes. -

Human Brain Organoids Reveal Accelerated Development of Cortical Neuron Classes As a Shared Feature of Autism Risk Genes
bioRxiv preprint doi: https://doi.org/10.1101/2020.11.10.376509; this version posted November 12, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Human brain organoids reveal accelerated development of cortical neuron classes as a shared feature of autism risk genes Bruna Paulsen1,2,†, Silvia Velasco1,2,†,#, Amanda J. Kedaigle1,2,3,†, Martina Pigoni1,2,†, Giorgia Quadrato4,5 Anthony Deo2,6,7,8, Xian Adiconis2,3, Ana Uzquiano1,2, Kwanho Kim1,2,3, Sean K. Simmons2,3, Kalliopi Tsafou2, Alex Albanese9, Rafaela Sartore1,2, Catherine Abbate1,2, Ashley Tucewicz1,2, Samantha Smith1,2, Kwanghun Chung9,10,11,12, Kasper Lage2,13, Aviv Regev3,14, Joshua Z. Levin2,3, Paola Arlotta1,2,# † These authors contributed equally to the work # Correspondence should be addressed to [email protected] and [email protected] 1 Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, MA 02138, USA 2 Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA 3 Klarman Cell Observatory, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA 4 Department of Stem Cell Biology and Regenerative Medicine, Keck School of Medicine, University of Southern California, Los Angeles, CA 90033, USA; 5 Eli and Edythe Broad CIRM Center for Regenerative Medicine and Stem Cell Research at the University of Southern California, Los Angeles, CA 90033, USA. 6 Department of Psychiatry,