Promiscuous Gene Expression in Thymic Epithelial Cells Is Regulated
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Role of Meprin Metalloproteases in Metastasis and Tumor Microenvironment
Cancer and Metastasis Reviews (2019) 38:347–356 https://doi.org/10.1007/s10555-019-09805-5 Role of meprin metalloproteases in metastasis and tumor microenvironment Florian Peters1 & Christoph Becker-Pauly1 Published online: 3 September 2019 # Springer Science+Business Media, LLC, part of Springer Nature 2019 Abstract A crucial step for tumor cell extravasation and metastasis is the migration through the extracellular matrix, which requires proteolytic activity. Hence, proteases, particularly matrix metalloproteases (MMPs), have been discussed as therapeutic targets and their inhibition should diminish tumor growth and metastasis. The metalloproteases meprin α and meprin β are highly abundant on intestinal enterocytes and their expression was associated with different stages of colorectal cancer. Due to their ability to cleave extracellular matrix (ECM) components, they were suggested as pro-tumorigenic enzymes. Additionally, both meprins were shown to have pro-inflammatory activity by cleaving cytokines and their receptors, which correlates with chronic intestinal inflammation and associated conditions. On the other hand, meprin β was identified as an essential enzyme for the detachment and renewal of the intestinal mucus, important to prevent bacterial overgrowth and infection. Considering this, it is hard to estimate whether high activity of meprins is generally detrimental or if these enzymes have also protective functions in certain cancer types. For instance, for colorectal cancer, patients with high meprin β expression in tumor tissue exhibit a better survival prognosis, which is completely different to prostate cancer. This demonstrates that the very same enzyme may have contrary effects on tumor initiation and growth, depending on its tissue and subcellular localization. Hence, precise knowledge about proteolytic enzymes is required to design the most efficient therapeutic options for cancer treatment. -

Metalloproteases Meprin Α and Meprin Β Are C- and N-Procollagen Proteinases Important for Collagen Assembly and Tensile Strength
Metalloproteases meprin α and meprin β are C- and N-procollagen proteinases important for collagen assembly and tensile strength Claudia Brodera, Philipp Arnoldb, Sandrine Vadon-Le Goffc, Moritz A. Konerdingd, Kerstin Bahrd, Stefan Müllere, Christopher M. Overallf, Judith S. Bondg, Tomas Koudelkah, Andreas Tholeyh, David J. S. Hulmesc, Catherine Moalic, and Christoph Becker-Paulya,1 aUnit for Degradomics of the Protease Web, Institute of Biochemistry, University of Kiel, 24118 Kiel, Germany; bInstitute of Zoology, Johannes Gutenberg University, 55128 Mainz, Germany; cTissue Biology and Therapeutic Engineering Unit, Centre National de la Recherche Scientifique/University of Lyon, Unité Mixte de Recherche 5305, Unité Mixte de Service 3444 Biosciences Gerland-Lyon Sud, 69367 Lyon Cedex 7, France; dInstitute of Functional and Clinical Anatomy, University Medical Center, Johannes Gutenberg University, 55128 Mainz, Germany; eDepartment of Gastroenterology, University of Bern, CH-3010 Bern, Switzerland; fCentre for Blood Research, University of British Columbia, Vancouver, BC, Canada V6T 1Z3; gDepartment of Biochemistry and Molecular Biology, Pennsylvania State University College of Medicine, Hershey, PA 17033; and hInstitute of Experimental Medicine, University of Kiel, 24118 Kiel, Germany Edited by Robert Huber, Max Planck Institute of Biochemistry, Planegg-Martinsried, Germany, and approved July 9, 2013 (received for review March 22, 2013) Type I fibrillar collagen is the most abundant protein in the human formation (22). A tight balance between synthesis and break- body, crucial for the formation and strength of bones, skin, and down of ECM is required for the function of all tissues, and tendon. Proteolytic enzymes are essential for initiation of the dysregulation leads to pathophysiological events, such as arthri- assembly of collagen fibrils by cleaving off the propeptides. -

Downloaded from the Protein Data Bank (PDB
bioRxiv preprint doi: https://doi.org/10.1101/2021.07.07.451411; this version posted July 7, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. CAT, AGTR2, L-SIGN and DC-SIGN are potential receptors for the entry of SARS-CoV-2 into human cells Dongjie Guo 1, 2, #, Ruifang Guo1, 2, #, Zhaoyang Li 1, 2, Yuyang Zhang 1, 2, Wei Zheng 3, Xiaoqiang Huang 3, Tursunjan Aziz 1, 2, Yang Zhang 3, 4, Lijun Liu 1, 2, * 1 College of Life and Health Sciences, Northeastern University, Shenyang, Liaoning, China 2 Key Laboratory of Data Analytics and Optimization for Smart Industry (Ministry of Education), Northeastern University, Shenyang, Liaoning, China 3 Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, USA 4 Department of Biological Chemistry, University of Michigan, Ann Arbor, USA * Corresponding author. College of Life and Health Sciences, Northeastern University, Shenyang, 110169, China. E-mail address: [email protected] (L. Liu) # These authors contributed equally to this work. 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.07.07.451411; this version posted July 7, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract Since December 2019, the COVID-19 caused by SARS-CoV-2 has been widely spread all over the world. -

MEP1A Allele for Meprin a Metalloprotease Is a Susceptibility Gene for Inflammatory Bowel Disease
ARTICLES nature publishing group See COMMENTARY page XX MEP1A allele for meprin A metalloprotease is a susceptibility gene for inflammatory bowel disease S B a n e r j e e 1 , 8 , B O n e d a 2 , 8 , L M Ya p 3 , 9 , D P J e w e l l 4 , G L M a t t e r s 1 , L R F it5 z p a , t r ic k F 6 S e , ib o l dE E 2 , S t e r c h i T A h m a d 7 , D L o t t a z 2,10 a n d J S B o n d 1 The MEP1A gene, located on human chromosome 6p (mouse chromosome 17) in a susceptibility region for inflammatory bowel disease (IBD), encodes the -subunit of metalloproteinase meprin A, which is expressed in the intestinal epithelium. This study shows a genetic association of MEP1A with IBD in a cohort of ulcerative colitis (UC) patients. There were four single-nucleotide polymorphisms in the coding region (P = 0.0012 – 0.04), and one in the 3 Ј -untranslated region ( P = 2 × 10 − 7 ) that displayed associations with UC. Moreover, meprin- mRNA was decreased in inflamed mucosa of IBD patients. Meprin- knockout mice exhibited a more severe intestinal injury and inflammation than their wild-type counterparts following oral administration of dextran sulfate sodium. Collectively, the data implicate MEP1A as a UC susceptibility gene and indicate that decreased meprin- expression is associated with intestinal inflammation in IBD patients and in a mouse experimental model of IBD. -

Functional and Structural Insights Into Astacin Metallopeptidases
Biol. Chem., Vol. 393, pp. 1027–1041, October 2012 • Copyright © by Walter de Gruyter • Berlin • Boston. DOI 10.1515/hsz-2012-0149 Review Functional and structural insights into astacin metallopeptidases F. Xavier Gomis-R ü th 1, *, Sergio Trillo-Muyo 1 Keywords: bone morphogenetic protein; catalytic domain; and Walter St ö cker 2, * meprin; metzincin; tolloid; zinc metallopeptidase. 1 Proteolysis Lab , Molecular Biology Institute of Barcelona, CSIC, Barcelona Science Park, Helix Building, c/Baldiri Reixac, 15-21, E-08028 Barcelona , Spain Introduction: a short historical background 2 Institute of Zoology , Cell and Matrix Biology, Johannes Gutenberg University, Johannes-von-M ü ller-Weg 6, The fi rst report on the digestive protease astacin from the D-55128 Mainz , Germany European freshwater crayfi sh, Astacus astacus L. – then termed ‘ crayfi sh small-molecule protease ’ or ‘ Astacus pro- * Corresponding authors tease ’ – dates back to the late 1960s (Sonneborn et al. , 1969 ). e-mail: [email protected]; [email protected] Protein sequencing by Zwilling and co-workers in the 1980s did not reveal homology to any other protein (Titani et al. , Abstract 1987 ). Shortly after, the enzyme was identifi ed as a zinc met- allopeptidase (St ö cker et al., 1988 ), and other family mem- The astacins are a family of multi-domain metallopepti- bers emerged. The fi rst of these was bone morphogenetic β dases with manifold functions in metabolism. They are protein 1 (BMP1), a protease co-purifi ed with TGF -like either secreted or membrane-anchored and are regulated growth factors termed bone morphogenetic proteins due by being synthesized as inactive zymogens and also by co- to their capacity to induce ectopic bone formation in mice localizing protein inhibitors. -

Origin and Diversification of Meprin Proteases
RESEARCH ARTICLE Origin and Diversification of Meprin Proteases Ignacio Marín* Instituto de Biomedicina de Valencia, Consejo Superior de Investigaciones Científicas, (IBV-CSIC), Valencia, Spain * [email protected] Abstract Meprins are astacin metalloproteases with a characteristic, easily recognizable structure, given that they are the only proteases with both MAM and MATH domains plus a trans- membrane region. So far assumed to be vertebrate-specific, it is shown here, using a com- bination of evolutionary and genomic analyses, that meprins originated before the urochordates/vertebrates split. In particular, three genes encoding structurally typical meprin proteins are arranged in tandem in the genome of the urochordate Ciona intestina- lis. Phylogenetic analyses showed that the protease and MATH domains present in the meprin-like proteins encoded by the Ciona genes are very similar in sequence to the OPEN ACCESS domains found in vertebrate meprins, which supports them having a common origin. While many vertebrates have the two canonical meprin-encoding genes orthologous to human Citation: Marín I (2015) Origin and Diversification of Meprin Proteases. PLoS ONE 10(8): e0135924. MEP1A and MEP1B (which respectively encode for the proteins known as meprin α and doi:10.1371/journal.pone.0135924 meprin β), a single gene has been found so far in the genome of the chondrichthyan fish Editor: Michael Schubert, Laboratoire de Biologie du Callorhinchus milii, and additional meprin-encoding genes are present in some species. Développement de Villefranche-sur-Mer, FRANCE Particularly, a group of bony fish species have genes encoding highly divergent meprins, Received: January 13, 2015 here named meprin-F. Genes encoding meprin-F proteins, derived from MEP1B genes, are abundant in some species, as the Amazon molly, Poecilia formosa, which has 7 of Accepted: July 28, 2015 them. -

Metalloproteases Meprin-Ɑ (MEP1A) Is a Prognostic Biomarker And
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Springer - Publisher Connector Wang et al. BMC Cancer (2016) 16:383 DOI 10.1186/s12885-016-2460-5 RESEARCH ARTICLE Open Access Metalloproteases meprin-ɑ (MEP1A) is a prognostic biomarker and promotes proliferation and invasion of colorectal cancer Xiao Wang1, Jian Chen1, Jingtao Wang1, Fudong Yu1, Senlin Zhao1, Yu Zhang2, Huamei Tang2 and Zhihai Peng1* Abstract Background: Meprin displays multiple functions in both health and disease, due in part to its broad proteolytic activity. In this report, we explored the clinical significance and functional relevance of the expression of meprin-ɑ (MEP1A) in colorectal cancer (CRC). Methods: The mRNA and protein expression levels of MEP1A in tumor specimens obtained from CRC patients was determined by quantitative real-time PCR and Western blot assay and comparatively paired with adjacent mucosa that presented as normal tissue. ShRNA was used to knock-down MEP1A expression in CRC cell-lines and the effects of dampened expression of MEP1A on the proliferation and invasion were determined by colony formation assays, Cell Counting Kit-8 assays and matrigel invasion assays. Moreover, nude mouse xenograft models were designed to investigate the same effect in vivo. In order to determine whether MEP1A expression correlated with CRC clinicopathologic factors and survival, immunohistochemical staining of a tissue microarray containing 88 paired CRC specimens was performed. Results: In CRC, enhanced expression of MEP1A was seen. Additionally, both in vitro and in vivo, CRC cellular proliferation and invasiveness was inhibited by dampened MEP1A expression. Several parameters were associated with enhanced MEP1A expression including tumor size (P = 0.023), staging of CRC by the American Joint Committee on Cancer (AJCC) (P = 0.024), and T (P = 0.032) and N stages (P = 0.001). -

Cov-2 Infection in the Small Intestine
Gastroenterology and Hepatology From Bed to Bench. ORIGINAL ARTICLE ©2020 RIGLD, Research Institute for Gastroenterology and Liver Diseases Investigating the human protein-host protein interactome of SARS- CoV-2 infection in the small intestine Mahmoud Khodadoost1, Zahra Niknam2, Masoumeh Farahani2, Mohammadreza Razzaghi3, Mohsen Norouzinia4 1Department of Traditional Medicine, School of Traditional Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran 2Proteomics Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran 3Laser Application in Medical Sciences Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran 4 Gastroenterology and Liver Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran ABSTRACT Aim: The present study aimed to identify human protein–host protein interactions of SARS-CoV-2 infection in the small intestine to discern the potential mechanisms and gain insights into the associated biomarkers and treatment strategies. Background: Deciphering the tissue and organ interactions of the SARS-CoV-2 infection can be important to discern the potential underlying mechanisms. In the present study, we investigated the human protein–host protein interactions in the small intestine. Methods: Public databases and published works were used to collect data related to small intestine tissue and SARS-CoV-2 infection. We constructed a human protein-protein interaction (PPI) network and showed interactions of host proteins in the small intestine. Associated modules, biological processes, functional pathways, regulatory transcription factors, disease ontology categories, and possible drug candidates for therapeutic targets were identified. Results: Thirteen primary protein neighbors were found for the SARS-CoV-2 receptor ACE2. ACE2 and its four partners were observed in a highly clustered module; moreover, 8 host proteins belonged to this module. -

The Structure, Function and Evolution of the Extracellular Matrix: a Systems-Level Analysis
The Structure, Function and Evolution of the Extracellular Matrix: A Systems-Level Analysis by Graham L. Cromar A thesis submitted in conformity with the requirements for the degree of Doctor of Philosophy Department of Molecular Genetics University of Toronto © Copyright by Graham L. Cromar 2014 ii The Structure, Function and Evolution of the Extracellular Matrix: A Systems-Level Analysis Graham L. Cromar Doctor of Philosophy Department of Molecular Genetics University of Toronto 2014 Abstract The extracellular matrix (ECM) is a three-dimensional meshwork of proteins, proteoglycans and polysaccharides imparting structure and mechanical stability to tissues. ECM dysfunction has been implicated in a number of debilitating conditions including cancer, atherosclerosis, asthma, fibrosis and arthritis. Identifying the components that comprise the ECM and understanding how they are organised within the matrix is key to uncovering its role in health and disease. This study defines a rigorous protocol for the rapid categorization of proteins comprising a biological system. Beginning with over 2000 candidate extracellular proteins, 357 core ECM genes and 524 functionally related (non-ECM) genes are identified. A network of high quality protein-protein interactions constructed from these core genes reveals the ECM is organised into biologically relevant functional modules whose components exhibit a mosaic of expression and conservation patterns. This suggests module innovations were widespread and evolved in parallel to convey tissue specific functionality on otherwise broadly expressed modules. Phylogenetic profiles of ECM proteins highlight components restricted and/or expanded in metazoans, vertebrates and mammals, indicating taxon-specific tissue innovations. Modules enriched for medical subject headings illustrate the potential for systems based analyses to predict new functional and disease associations on the basis of network topology. -

Common Genetic Variation at the IL1RL1 Locus Regulates IL- 33/ST2 Signaling
Common genetic variation at the IL1RL1 locus regulates IL- 33/ST2 signaling Jennifer E. Ho, … , Richard T. Lee, Thomas J. Wang J Clin Invest. 2013;123(10):4208-4218. https://doi.org/10.1172/JCI67119. Research Article Immunology The suppression of tumorigenicity 2/IL-33 (ST2/IL-33) pathway has been implicated in several immune and inflammatory diseases. ST2 is produced as 2 isoforms. The membrane-bound isoform (ST2L) induces an immune response when bound to its ligand, IL-33. The other isoform is a soluble protein (sST2) that is thought to be a decoy receptor for IL-33 signaling. Elevated sST2 levels in serum are associated with an increased risk for cardiovascular disease. We investigated the determinants of sST2 plasma concentrations in 2,991 Framingham Offspring Cohort participants. While clinical and environmental factors explained some variation in sST2 levels, much of the variation in sST2 production was driven by genetic factors. In a genome-wide association study (GWAS), multiple SNPs within IL1RL1 (the gene encoding ST2) demonstrated associations with sST2 concentrations. Five missense variants of IL1RL1 correlated with higher sST2 levels in the GWAS and mapped to the intracellular domain of ST2, which is absent in sST2. In a cell culture model, IL1RL1 missense variants increased sST2 expression by inducing IL-33 expression and enhancing IL-33 responsiveness (via ST2L). Our data suggest that genetic variation in IL1RL1 can result in increased levels of sST2 and alter immune and inflammatory signaling through the ST2/IL-33 pathway. Find the latest version: https://jci.me/67119/pdf Research article Common genetic variation at the IL1RL1 locus regulates IL-33/ST2 signaling Jennifer E. -
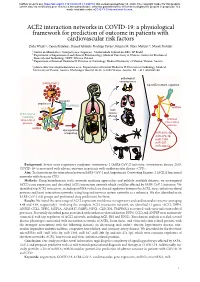
ACE2 Interaction Networks in COVID-19
bioRxiv preprint doi: https://doi.org/10.1101/2020.05.13.094714; this version posted May 14, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. ACE2 interaction networks in COVID-19: a physiological framework for prediction of outcome in patients with cardiovascular risk factors Zofia Wicik1,2, Ceren Eyileten2, Daniel Jakubik2, Rodrigo Pavão1, Jolanta M. Siller-Matula2,3*, Marek Postula2 ¹ Centro de Matemática, Computação e Cognição - Universidade Federal do ABC, SP, Brazil ² Department of Experimental and Clinical Pharmacology, Medical University of Warsaw, Center for Preclinical Research and Technology CEPT, Warsaw, Poland ³ Department of Internal Medicine II, Division of Cardiology, Medical University of Vienna, Vienna, Austria * [email protected]; Department of Internal Medicine II, Division of Cardiology, Medical University of Vienna, Austria. Waehringer Guertel 18-20, A-1090 Vienna, Austria. Tel. +43 1 4040046140 pathological consequences SARS-CoV-2 top ACE2 network regulators activation attachment TMPRSS2 miRNA ACE2 main ACE2 network tissues containing affected virus-related activated proteins virus-related proteins miRNA dysregulation of signaling miR-302c-5p miR-27a-3p miR-1305 miR-587 miR-26b-5p Background: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection (coronavirus disease 2019; COVID-19) is associated with adverse outcome in patients with cardiovascular disease (CVD). Aim: To characterize the interaction between SARS-CoV-2 and Angiotensin Converting Enzyme 2 (ACE2) functional networks with focus on CVD. -

Research Article in Silico Analysis of High-Risk Missense Variants in Human ACE2 Gene and Susceptibility to SARS-Cov-2 Infection
Hindawi BioMed Research International Volume 2021, Article ID 6685840, 10 pages https://doi.org/10.1155/2021/6685840 Research Article In Silico Analysis of High-Risk Missense Variants in Human ACE2 Gene and Susceptibility to SARS-CoV-2 Infection Asmae Saih ,1,2 Hana Baba,1,2 Meryem Bouqdayr,1,2 Hassan Ghazal,3,4 Salsabil Hamdi,5 Anass Kettani,2 and Lahcen Wakrim1 1Virology Unit, Immunovirology Laboratory, Institut Pasteur du Maroc, 20360 Casablanca, Morocco 2Laboratory of Biology and Health, URAC 34, Faculty of Sciences Ben M’Sik Hassan II University of Casablanca, Morocco 3National Center for Scientific Technical Research (CNRST), Rabat 10102, Morocco 4Department of Fundamental Sciences, School of Medicine, Mohammed VI University of Health Sciences, Casablanca, Morocco 5Environmental Health Laboratory, Institut Pasteur du Maroc, 20360 Casablanca, Morocco Correspondence should be addressed to Asmae Saih; [email protected] Received 1 January 2021; Revised 8 March 2021; Accepted 18 March 2021; Published 9 April 2021 Academic Editor: R. K. Tripathy Copyright © 2021 Asmae Saih et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. SARS-CoV-2 coronavirus uses for entry to human host cells a SARS-CoV receptor of the angiotensin-converting enzyme (ACE2) that catalyzes the conversion of angiotensin II into angiotensin (1-7). To understand the effect of ACE2 missense variants on protein structure, stability, and function, various bioinformatics tools were used including SIFT, PANTHER, PROVEAN, PolyPhen2.0, I. Mutant Suite, MUpro, SWISS-MODEL, Project HOPE, ModPred, QMEAN, ConSurf, and STRING.