Precursors, Causes, Consequences—The Choanoflagellate/Sponge Transition, Neurogen- Phil
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Buzzle – Zoology Terms – Glossary of Biology Terms and Definitions Http
Buzzle – Zoology Terms – Glossary of Biology Terms and Definitions http://www.buzzle.com/articles/biology-terms-glossary-of-biology-terms-and- definitions.html#ZoologyGlossary Biology is the branch of science concerned with the study of life: structure, growth, functioning and evolution of living things. This discipline of science comprises three sub-disciplines that are botany (study of plants), Zoology (study of animals) and Microbiology (study of microorganisms). This vast subject of science involves the usage of myriads of biology terms, which are essential to be comprehended correctly. People involved in the science field encounter innumerable jargons during their study, research or work. Moreover, since science is a part of everybody's life, it is something that is important to all individuals. A Abdomen: Abdomen in mammals is the portion of the body which is located below the rib cage, and in arthropods below the thorax. It is the cavity that contains stomach, intestines, etc. Abscission: Abscission is a process of shedding or separating part of an organism from the rest of it. Common examples are that of, plant parts like leaves, fruits, flowers and bark being separated from the plant. Accidental: Accidental refers to the occurrences or existence of all those species that would not be found in a particular region under normal circumstances. Acclimation: Acclimation refers to the morphological and/or physiological changes experienced by various organisms to adapt or accustom themselves to a new climate or environment. Active Transport: The movement of cellular substances like ions or molecules by traveling across the membrane, towards a higher level of concentration while consuming energy. -

The Closest Unicellular Relatives of Animals
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Current Biology, Vol. 12, 1773–1778, October 15, 2002, 2002 Elsevier Science Ltd. All rights reserved. PII S0960-9822(02)01187-9 The Closest Unicellular Relatives of Animals B.F. Lang,1,2 C. O’Kelly,1,3 T. Nerad,4 M.W. Gray,1,5 Results and Discussion and G. Burger1,2,6 1The Canadian Institute for Advanced Research The evolution of the Metazoa from single-celled protists Program in Evolutionary Biology is an issue that has intrigued biologists for more than a 2 De´ partement de Biochimie century. Early morphological and more recent ultra- Universite´ de Montre´ al structural and molecular studies have converged in sup- Succursale Centre-Ville porting the now widely accepted view that animals are Montre´ al, Que´ bec H3C 3J7 related to Fungi, choanoflagellates, and ichthyosporean Canada protists. However, controversy persists as to the spe- 3 Bigelow Laboratory for Ocean Sciences cific evolutionary relationships among these major P.O. Box 475 groups. This uncertainty is reflected in the plethora of 180 McKown Point Road published molecular phylogenies that propose virtually West Boothbay Harbor, Maine 04575 all of the possible alternative tree topologies involving 4 American Type Culture Collection Choanoflagellata, Fungi, Ichthyosporea, and Metazoa. 10801 University Boulevard For example, a monophyletic MetazoaϩChoanoflagel- Manassas, Virginia 20110 lata group has been suggested on the basis of small 5 Department of Biochemistry subunit (SSU) rDNA sequences [1, 6, 7]. Other studies and Molecular Biology using the same sequences have allied Choanoflagellata Dalhousie University with the Fungi [8], placed Choanoflagellata prior to the Halifax, Nova Scotia B3H 4H7 divergence of animals and Fungi [9], or even placed Canada them prior to the divergence of green algae and land plants [10]. -

Multigene Eukaryote Phylogeny Reveals the Likely Protozoan Ancestors of Opis- Thokonts (Animals, Fungi, Choanozoans) and Amoebozoa
Accepted Manuscript Multigene eukaryote phylogeny reveals the likely protozoan ancestors of opis- thokonts (animals, fungi, choanozoans) and Amoebozoa Thomas Cavalier-Smith, Ema E. Chao, Elizabeth A. Snell, Cédric Berney, Anna Maria Fiore-Donno, Rhodri Lewis PII: S1055-7903(14)00279-6 DOI: http://dx.doi.org/10.1016/j.ympev.2014.08.012 Reference: YMPEV 4996 To appear in: Molecular Phylogenetics and Evolution Received Date: 24 January 2014 Revised Date: 2 August 2014 Accepted Date: 11 August 2014 Please cite this article as: Cavalier-Smith, T., Chao, E.E., Snell, E.A., Berney, C., Fiore-Donno, A.M., Lewis, R., Multigene eukaryote phylogeny reveals the likely protozoan ancestors of opisthokonts (animals, fungi, choanozoans) and Amoebozoa, Molecular Phylogenetics and Evolution (2014), doi: http://dx.doi.org/10.1016/ j.ympev.2014.08.012 This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain. 1 1 Multigene eukaryote phylogeny reveals the likely protozoan ancestors of opisthokonts 2 (animals, fungi, choanozoans) and Amoebozoa 3 4 Thomas Cavalier-Smith1, Ema E. Chao1, Elizabeth A. Snell1, Cédric Berney1,2, Anna Maria 5 Fiore-Donno1,3, and Rhodri Lewis1 6 7 1Department of Zoology, University of Oxford, South Parks Road, Oxford OX1 3PS, UK. -
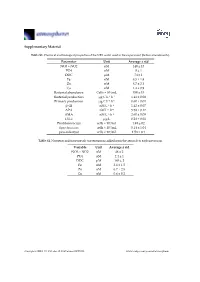
Supplementary Material Parameter Unit Average ± Std NO3 + NO2 Nm
Supplementary Material Table S1. Chemical and biological properties of the NRS water used in the experiment (before amendments). Parameter Unit Average ± std NO3 + NO2 nM 140 ± 13 PO4 nM 8 ± 1 DOC μM 74 ± 1 Fe nM 8.5 ± 1.8 Zn nM 8.7 ± 2.1 Cu nM 1.4 ± 0.9 Bacterial abundance Cells × 104/mL 350 ± 15 Bacterial production μg C L−1 h−1 1.41 ± 0.08 Primary production μg C L−1 h−1 0.60 ± 0.01 β-Gl nM L−1 h−1 1.42 ± 0.07 APA nM L−1 h−1 5.58 ± 0.17 AMA nM L−1·h−1 2.60 ± 0.09 Chl-a μg/L 0.28 ± 0.01 Prochlorococcus cells × 104/mL 1.49 ± 02 Synechococcus cells × 104/mL 5.14 ± 1.04 pico-eukaryot cells × 103/mL 1.58 × 0.1 Table S2. Nutrients and trace metals concentrations added from the aerosols to each mesocosm. Variable Unit Average ± std NO3 + NO2 nM 48 ± 2 PO4 nM 2.4 ± 1 DOC μM 165 ± 2 Fe nM 2.6 ± 1.5 Zn nM 6.7 ± 2.5 Cu nM 0.6 ± 0.2 Atmosphere 2019, 10, 358; doi:10.3390/atmos10070358 www.mdpi.com/journal/atmosphere Atmosphere 2019, 10, 358 2 of 6 Table S3. ANOVA test results between control, ‘UV-treated’ and ‘live-dust’ treatments at 20 h or 44 h, with significantly different values shown in bold. ANOVA df Sum Sq Mean Sq F Value p-value Chl-a 20 H 2, 6 0.03, 0.02 0.02, 0 4.52 0.0634 44 H 2, 6 0.02, 0 0.01, 0 23.13 0.002 Synechococcus Abundance 20 H 2, 7 8.23 × 107, 4.11 × 107 4.11 × 107, 4.51 × 107 0.91 0.4509 44 H 2, 7 5.31 × 108, 6.97 × 107 2.65 × 108, 1.16 × 107 22.84 0.0016 Prochlorococcus Abundance 20 H 2, 8 4.22 × 107, 2.11 × 107 2.11 × 107, 2.71 × 106 7.77 0.0216 44 H 2, 8 9.02 × 107, 1.47 × 107 4.51 × 107, 2.45 × 106 18.38 0.0028 Pico-eukaryote -

Predatory Flagellates – the New Recently Discovered Deep Branches of the Eukaryotic Tree and Their Evolutionary and Ecological Significance
Protistology 14 (1), 15–22 (2020) Protistology Predatory flagellates – the new recently discovered deep branches of the eukaryotic tree and their evolutionary and ecological significance Denis V. Tikhonenkov Papanin Institute for Biology of Inland Waters, Russian Academy of Sciences, Borok, 152742, Russia | Submitted March 20, 2020 | Accepted April 6, 2020 | Summary Predatory protists are poorly studied, although they are often representing important deep-branching evolutionary lineages and new eukaryotic supergroups. This short review/opinion paper is inspired by the recent discoveries of various predatory flagellates, which form sister groups of the giant eukaryotic clusters on phylogenetic trees, and illustrate an ancestral state of one or another supergroup of eukaryotes. Here we discuss their evolutionary and ecological relevance and show that the study of such protists may be essential in addressing previously puzzling evolutionary problems, such as the origin of multicellular animals, the plastid spread trajectory, origins of photosynthesis and parasitism, evolution of mitochondrial genomes. Key words: evolution of eukaryotes, heterotrophic flagellates, mitochondrial genome, origin of animals, photosynthesis, predatory protists, tree of life Predatory flagellates and diversity of eu- of the hidden diversity of protists (Moon-van der karyotes Staay et al., 2000; López-García et al., 2001; Edg- comb et al., 2002; Massana et al., 2004; Richards The well-studied multicellular animals, plants and Bass, 2005; Tarbe et al., 2011; de Vargas et al., and fungi immediately come to mind when we hear 2015). In particular, several prevailing and very abun- the term “eukaryotes”. However, these groups of dant ribogroups such as MALV, MAST, MAOP, organisms represent a minority in the real diversity MAFO (marine alveolates, stramenopiles, opistho- of evolutionary lineages of eukaryotes. -

Protist Phylogeny and the High-Level Classification of Protozoa
Europ. J. Protistol. 39, 338–348 (2003) © Urban & Fischer Verlag http://www.urbanfischer.de/journals/ejp Protist phylogeny and the high-level classification of Protozoa Thomas Cavalier-Smith Department of Zoology, University of Oxford, South Parks Road, Oxford, OX1 3PS, UK; E-mail: [email protected] Received 1 September 2003; 29 September 2003. Accepted: 29 September 2003 Protist large-scale phylogeny is briefly reviewed and a revised higher classification of the kingdom Pro- tozoa into 11 phyla presented. Complementary gene fusions reveal a fundamental bifurcation among eu- karyotes between two major clades: the ancestrally uniciliate (often unicentriolar) unikonts and the an- cestrally biciliate bikonts, which undergo ciliary transformation by converting a younger anterior cilium into a dissimilar older posterior cilium. Unikonts comprise the ancestrally unikont protozoan phylum Amoebozoa and the opisthokonts (kingdom Animalia, phylum Choanozoa, their sisters or ancestors; and kingdom Fungi). They share a derived triple-gene fusion, absent from bikonts. Bikonts contrastingly share a derived gene fusion between dihydrofolate reductase and thymidylate synthase and include plants and all other protists, comprising the protozoan infrakingdoms Rhizaria [phyla Cercozoa and Re- taria (Radiozoa, Foraminifera)] and Excavata (phyla Loukozoa, Metamonada, Euglenozoa, Percolozoa), plus the kingdom Plantae [Viridaeplantae, Rhodophyta (sisters); Glaucophyta], the chromalveolate clade, and the protozoan phylum Apusozoa (Thecomonadea, Diphylleida). Chromalveolates comprise kingdom Chromista (Cryptista, Heterokonta, Haptophyta) and the protozoan infrakingdom Alveolata [phyla Cilio- phora and Miozoa (= Protalveolata, Dinozoa, Apicomplexa)], which diverged from a common ancestor that enslaved a red alga and evolved novel plastid protein-targeting machinery via the host rough ER and the enslaved algal plasma membrane (periplastid membrane). -

S41467-021-25308-W.Pdf
ARTICLE https://doi.org/10.1038/s41467-021-25308-w OPEN Phylogenomics of a new fungal phylum reveals multiple waves of reductive evolution across Holomycota ✉ ✉ Luis Javier Galindo 1 , Purificación López-García 1, Guifré Torruella1, Sergey Karpov2,3 & David Moreira 1 Compared to multicellular fungi and unicellular yeasts, unicellular fungi with free-living fla- gellated stages (zoospores) remain poorly known and their phylogenetic position is often 1234567890():,; unresolved. Recently, rRNA gene phylogenetic analyses of two atypical parasitic fungi with amoeboid zoospores and long kinetosomes, the sanchytrids Amoeboradix gromovi and San- chytrium tribonematis, showed that they formed a monophyletic group without close affinity with known fungal clades. Here, we sequence single-cell genomes for both species to assess their phylogenetic position and evolution. Phylogenomic analyses using different protein datasets and a comprehensive taxon sampling result in an almost fully-resolved fungal tree, with Chytridiomycota as sister to all other fungi, and sanchytrids forming a well-supported, fast-evolving clade sister to Blastocladiomycota. Comparative genomic analyses across fungi and their allies (Holomycota) reveal an atypically reduced metabolic repertoire for sanchy- trids. We infer three main independent flagellum losses from the distribution of over 60 flagellum-specific proteins across Holomycota. Based on sanchytrids’ phylogenetic position and unique traits, we propose the designation of a novel phylum, Sanchytriomycota. In addition, our results indicate that most of the hyphal morphogenesis gene repertoire of multicellular fungi had already evolved in early holomycotan lineages. 1 Ecologie Systématique Evolution, CNRS, Université Paris-Saclay, AgroParisTech, Orsay, France. 2 Zoological Institute, Russian Academy of Sciences, St. ✉ Petersburg, Russia. 3 St. -

Introns and Alternative Splicing in Choanoflagellates
Introns and alternative splicing in choanoflagellates By Marjorie Wright Westbrook A dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Molecular and Cell Biology in the Graduate Division of the University of California, Berkeley Committee in charge: Professor Nicole King Professor Donald C. Rio Professor G. Steven Martin Professor Steven E. Brenner Fall 2011 Introns and alternative splicing in choanoflagellates ©2011 by Marjorie Wright Westbrook Abstract Introns and alternative splicing in choanoflagellates By Marjorie Wright Westbrook Doctor of Philosophy in Molecular and Cell Biology University of California, Berkeley Professor Nicole King, Chair The first organisms to evolve were unicellular, and the vast majority of life has remained so for billions of years. Complex forms of multicellularity, requiring increased levels of cell adhesion, cell signaling and gene regulation, have evolved in only a few eukaryotic lineages [1, 2]. The comparison of genomes from choanoflagellates, the closest relatives of metazoans, with genomes from metazoans may reveal genomic changes underlying metazoan origins. I used this approach to investigate the evolution of introns during the origin of metazoans. By analyzing the genome of the first choanoflagellate to be sequenced, Monosiga brevicollis, I found that its intron density rivals that of genes in intron-rich metazoans [3]. Many intron positions are conserved between choanoflagellates and metazoans, implying that their shared unicellular ancestor was also intron-rich. In my analysis of the M. brevicollis genome, I made the unexpected discovery that, unlike most choanoflagellate genes, the longest genes contain relatively few introns. Indeed, one M. brevicollis gene contains the longest stretch of intron-free coding sequence known to date. -

Diversity and Distribution of Unicellular Opisthokonts Along the European Coast Analyzed Using High-Throughput Sequencing
Diversity and distribution of unicellular opisthokonts along the European coast analyzed using high-throughput sequencing Javier del Campo1,2,*, Diego Mallo3, Ramon Massana4, Colomban de Vargas5,6, Thomas A. Richards7,8, and Iñaki Ruiz-Trillo1,9,10 1Institut de Biologia Evolutiva (CSIC-UPF), Passeig Marítim de la Barceloneta 37-49, 08003 Barcelona, Catalonia, Spain. 3Department of Biochemistry, Genetics and Immunology, University of Vigo, Vigo, Galicia, Spain 4Department of Marine Biology and Oceanography, Institut de Ciències del Mar (CSIC), Barcelona, Catalonia, Spain 5CNRS, UMR 7144, Adaptation et Diversité en Milieu Marin, Station Biologique de Roscoff, Roscoff, France 6UPMC Univ. Paris 06, UMR 7144, Station Biologique de Roscoff, Roscoff, France 7Geoffrey Pope Building, Biosciences, College of Life and Environmental Sciences, University of Exeter, Exeter, UK 8Canadian Institute for Advanced Research, CIFAR Program in Integrated Microbial Biodiversity 9Departament de Genètica, Universitat de Barcelona, Barcelona, Catalonia, Spain 10Institució Catalana de Recerca i Estudis Avançats (ICREA), Barcelona, Catalonia, Spain Summary The opisthokonts are one of the major super-groups of eukaryotes. It comprises two major clades: 1) the Metazoa and their unicellular relatives and 2) the Fungi and their unicellular relatives. There is, however, little knowledge of the role of opisthokont microbes in many natural environments, especially among non-metazoan and non-fungal opisthokonts. Here we begin to address this gap by analyzing high throughput 18S rDNA and 18S rRNA sequencing data from different European coastal sites, sampled at different size fractions and depths. In particular, we analyze the diversity and abundance of choanoflagellates, filastereans, ichthyosporeans, nucleariids, corallochytreans and their related lineages. Our results show the great diversity of choanoflagellates in coastal waters as well as a relevant role of the ichthyosporeans and the uncultured marine opisthokonts (MAOP). -

Controlled Sampling of Ribosomally Active Protistan Diversity in Sediment-Surface Layers Identifies Putative Players in the Marine Carbon Sink
The ISME Journal (2020) 14:984–998 https://doi.org/10.1038/s41396-019-0581-y ARTICLE Controlled sampling of ribosomally active protistan diversity in sediment-surface layers identifies putative players in the marine carbon sink 1,2 1 1 3 3 Raquel Rodríguez-Martínez ● Guy Leonard ● David S. Milner ● Sebastian Sudek ● Mike Conway ● 1 1 4,5 6 7 Karen Moore ● Theresa Hudson ● Frédéric Mahé ● Patrick J. Keeling ● Alyson E. Santoro ● 3,8 1,9 Alexandra Z. Worden ● Thomas A. Richards Received: 6 October 2019 / Revised: 4 December 2019 / Accepted: 17 December 2019 / Published online: 9 January 2020 © The Author(s) 2020. This article is published with open access Abstract Marine sediments are one of the largest carbon reservoir on Earth, yet the microbial communities, especially the eukaryotes, that drive these ecosystems are poorly characterised. Here, we report implementation of a sampling system that enables injection of reagents into sediments at depth, allowing for preservation of RNA in situ. Using the RNA templates recovered, we investigate the ‘ribosomally active’ eukaryotic diversity present in sediments close to the water/sediment interface. We 1234567890();,: 1234567890();,: demonstrate that in situ preservation leads to recovery of a significantly altered community profile. Using SSU rRNA amplicon sequencing, we investigated the community structure in these environments, demonstrating a wide diversity and high relative abundance of stramenopiles and alveolates, specifically: Bacillariophyta (diatoms), labyrinthulomycetes and ciliates. The identification of abundant diatom rRNA molecules is consistent with microscopy-based studies, but demonstrates that these algae can also be exported to the sediment as active cells as opposed to dead forms. -

Microbial Eukaryotes in Oil Sands Environments: Heterotrophs in the Spotlight
microorganisms Review Microbial Eukaryotes in Oil Sands Environments: Heterotrophs in the Spotlight Elisabeth Richardson 1,* and Joel B. Dacks 2,* 1 Department of Biological Sciences, University of Alberta, Edmonton, AB T6G 2E9, Canada 2 Division of Infectious Disease, Department of Medicine, University of Alberta, Edmonton, AB T6G 2G3, Canada * Correspondence: [email protected] (E.R.); [email protected] (J.B.D.); Tel.: +1-780-248-1493 (J.B.D.) Received: 6 May 2019; Accepted: 14 June 2019; Published: 19 June 2019 Abstract: Hydrocarbon extraction and exploitation is a global, trillion-dollar industry. However, for decades it has also been known that fossil fuel usage is environmentally detrimental; the burning of hydrocarbons results in climate change, and environmental damage during extraction and transport can also occur. Substantial global efforts into mitigating this environmental disruption are underway. The global petroleum industry is moving more and more into exploiting unconventional oil reserves, such as oil sands and shale oil. The Albertan oil sands are one example of unconventional oil reserves; this mixture of sand and heavy bitumen lying under the boreal forest of Northern Alberta represent one of the world’s largest hydrocarbon reserves, but extraction also requires the disturbance of a delicate northern ecosystem. Considerable effort is being made by various stakeholders to mitigate environmental impact and reclaim anthropogenically disturbed environments associated with oil sand extraction. In this review, we discuss the eukaryotic microbial communities associated with the boreal ecosystem and how this is affected by hydrocarbon extraction, with a particular emphasis on the reclamation of tailings ponds, where oil sands extraction waste is stored. -

THE STORY of ALCYONIUM: from HALCYON BIRDS to ZOOPHYTES E Robson
THE STORY OF ALCYONIUM: FROM HALCYON BIRDS TO ZOOPHYTES E Robson To cite this version: E Robson. THE STORY OF ALCYONIUM: FROM HALCYON BIRDS TO ZOOPHYTES. Vie et Milieu / Life & Environment, Observatoire Océanologique - Laboratoire Arago, 2002, pp.217-222. hal-03198950 HAL Id: hal-03198950 https://hal.sorbonne-universite.fr/hal-03198950 Submitted on 15 Apr 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. VIE MILIEU, 2002, 52 (4) : 217-222 From Marine Ecology to Developmental Biology In Honour of Pierre Tardent (1927-1997) THE STORY OF ALCYONIUM: FROM HALCYON BIRDS TO ZOOPHYTES E.A. ROBSON School of Animal & Microbial Sciences, The University of Reading, P.O. Box 228, Reading RG6 6AJ, UK ALCYONIUM ABSTRACT. - The name of the soft coral Alcyonium is derived from the ancient ALCYONARIA Greek myth of Ceyx and Alcyone. They were transformed into halcyon birds whose HALCYONS floating nests, empty after the young hatched, were mistakenly identified with Me- ZOOPHYTE MYTHOLOGY diterranean flotsam including the remains of sessile colonies or seaweeds detached HISTORY by wave action and so variously referred to as alcyoniums. During the eighteenth century ground-breaking research on Hydra by Trembley (1744) was followed by précise descriptions of hydroids and of Alcyonium digitatum (L.) (de Jussieu 1742 and especially Ellis 1755) and the récognition that ail "zoophytes" were animais.