Avant-Propos Le Format De Présentation De Cette Thèse Correspond À Une Recommandation À La Spécialité
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

National Center for Toxicological Research
National Center for Toxicological Research Annual Report Research Accomplishments and Plans FY 2015 – FY 2016 Page 0 of 193 Table of Contents Preface – William Slikker, Jr., Ph.D. ................................................................................... 3 NCTR Vision ......................................................................................................................... 7 NCTR Mission ...................................................................................................................... 7 NCTR Strategic Plan ............................................................................................................ 7 NCTR Organizational Structure .......................................................................................... 8 NCTR Location and Facilities .............................................................................................. 9 NCTR Advances Research Through Outreach and Collaboration ................................... 10 NCTR Global Outreach and Training Activities ............................................................... 12 Global Summit on Regulatory Science .................................................................................................12 Training Activities .................................................................................................................................14 NCTR Scientists – Leaders in the Research Community .................................................. 15 Science Advisory Board ................................................................................................... -

A Screening-Based Approach to Circumvent Tumor Microenvironment
JBXXXX10.1177/1087057113501081Journal of Biomolecular ScreeningSingh et al. 501081research-article2013 Original Research Journal of Biomolecular Screening 2014, Vol 19(1) 158 –167 A Screening-Based Approach to © 2013 Society for Laboratory Automation and Screening DOI: 10.1177/1087057113501081 Circumvent Tumor Microenvironment- jbx.sagepub.com Driven Intrinsic Resistance to BCR-ABL+ Inhibitors in Ph+ Acute Lymphoblastic Leukemia Harpreet Singh1,2, Anang A. Shelat3, Amandeep Singh4, Nidal Boulos1, Richard T. Williams1,2*, and R. Kiplin Guy2,3 Abstract Signaling by the BCR-ABL fusion kinase drives Philadelphia chromosome–positive acute lymphoblastic leukemia (Ph+ ALL) and chronic myelogenous leukemia (CML). Despite their clinical activity in many patients with CML, the BCR-ABL kinase inhibitors (BCR-ABL-KIs) imatinib, dasatinib, and nilotinib provide only transient leukemia reduction in patients with Ph+ ALL. While host-derived growth factors in the leukemia microenvironment have been invoked to explain this drug resistance, their relative contribution remains uncertain. Using genetically defined murine Ph+ ALL cells, we identified interleukin 7 (IL-7) as the dominant host factor that attenuates response to BCR-ABL-KIs. To identify potential combination drugs that could overcome this IL-7–dependent BCR-ABL-KI–resistant phenotype, we screened a small-molecule library including Food and Drug Administration–approved drugs. Among the validated hits, the well-tolerated antimalarial drug dihydroartemisinin (DHA) displayed potent activity in vitro and modest in vivo monotherapy activity against engineered murine BCR-ABL-KI–resistant Ph+ ALL. Strikingly, cotreatment with DHA and dasatinib in vivo strongly reduced primary leukemia burden and improved long-term survival in a murine model that faithfully captures the BCR-ABL-KI–resistant phenotype of human Ph+ ALL. -

Rapport 2008
rapport 2008 Reseptregisteret 2004-2007 The Norwegian Prescription Database 2004-2007 Marit Rønning Christian Lie Berg Kari Furu Irene Litleskare Solveig Sakshaug Hanne Strøm Rapport 2008 Nasjonalt folkehelseinstitutt/ The Norwegian Institute of Public Health Tittel/Title: Reseptregisteret 2004-2007 The Norwegian Prescription Database 2004-2007 Redaktør/Editor: Marit Rønning Forfattere/Authors: Christian Lie Berg Kari Furu Irene Litleskare Marit Rønning Solveig Sakshaug Hanne Strøm Publisert av/Published by: Nasjonalt folkehelseinstitutt Postboks 4404 Nydalen NO-0403 Norway Tel: + 47 21 07 70 00 E-mail: [email protected] www.fhi.no Design: Per Kristian Svendsen Layout: Grete Søimer Acknowledgement: Julie D.W. Johansen (English version) Forsideillustrasjon/Front page illustration: Colourbox.com Trykk/Print: Nordberg Trykk AS Opplag/ Number printed: 1200 Bestilling/Order: [email protected] Fax: +47-21 07 81 05 Tel: +47-21 07 82 00 ISSN: 0332-6535 ISBN: 978-82-8082-252-9 trykt utgave/printed version ISBN: 978-82-8082-253-6 elektronisk utgave/electronic version 2 Rapport 2008 • Folkehelseinstituttet Forord Bruken av legemidler i befolkningen er økende. En viktig målsetting for norsk legemiddelpolitikk er rasjonell legemiddelbruk. En forutsetning for arbeidet med å optimalisere legemiddelbruken i befolkningen er kunnskap om hvilke legemidler som brukes, hvem som bruker legemidlene og hvordan de brukes. For å få bedre kunnskap på dette området, vedtok Stortinget i desember 2002 å etablere et nasjonalt reseptbasert legemiddelregister (Reseptregisteret). Oppgaven med å etablere registeret ble gitt til Folkehelseinstituttet som fra 1. januar 2004 har mottatt månedlige opplysninger fra alle apotek om utlevering av legemidler til pasienter, leger og institusjoner. Denne rapporten er første utgave i en planlagt årlig statistikk fra Reseptregisteret. -

Step-By-Step Guide to Better Laboratory Management Practices
Step-by-Step Guide to Better Laboratory Management Practices Prepared by The Washington State Department of Ecology Hazardous Waste and Toxics Reduction Program Publication No. 97- 431 Revised January 2003 Printed on recycled paper For additional copies of this document, contact: Department of Ecology Publications Distribution Center PO Box 47600 Olympia, WA 98504-7600 (360) 407-7472 or 1 (800) 633-7585 or contact your regional office: Department of Ecology’s Regional Offices (425) 649-7000 (509) 575-2490 (509) 329-3400 (360) 407-6300 The Department of Ecology is an equal opportunity agency and does not discriminate on the basis of race, creed, color, disability, age, religion, national origin, sex, marital status, disabled veteran’s status, Vietnam Era veteran’s status or sexual orientation. If you have special accommodation needs, or require this document in an alternate format, contact the Hazardous Waste and Toxics Reduction Program at (360)407-6700 (voice) or 711 or (800) 833-6388 (TTY). Table of Contents Introduction ....................................................................................................................................iii Section 1 Laboratory Hazardous Waste Management ...........................................................1 Designating Dangerous Waste................................................................................................1 Counting Wastes .......................................................................................................................8 Treatment by Generator...........................................................................................................12 -

Chlorhexidine Bathing in a Tertiary Care Neonatal Intensive Care Unit
Chlorhexidine Bathing in a Tertiary Care Neonatal Intensive Care Unit: Impact on Central Line–Associated Bloodstream Infections Author(s): Caroline Quach, MD, MSc; Aaron M. Milstone, MD, MHS; Chantal Perpête, RN, LSH, LSHH; Mario Bonenfant, RN; Dorothy L. Moore, MD, PhD; Therese Perreault, MD Source: Infection Control and Hospital Epidemiology, Vol. 35, No. 2 (February 2014), pp. 158- 163 Published by: The University of Chicago Press on behalf of The Society for Healthcare Epidemiology of America Stable URL: http://www.jstor.org/stable/10.1086/674862 . Accessed: 24/01/2014 14:25 Your use of the JSTOR archive indicates your acceptance of the Terms & Conditions of Use, available at . http://www.jstor.org/page/info/about/policies/terms.jsp . JSTOR is a not-for-profit service that helps scholars, researchers, and students discover, use, and build upon a wide range of content in a trusted digital archive. We use information technology and tools to increase productivity and facilitate new forms of scholarship. For more information about JSTOR, please contact [email protected]. The University of Chicago Press and The Society for Healthcare Epidemiology of America are collaborating with JSTOR to digitize, preserve and extend access to Infection Control and Hospital Epidemiology. http://www.jstor.org This content downloaded from 209.172.182.131 on Fri, 24 Jan 2014 14:25:10 PM All use subject to JSTOR Terms and Conditions infection control and hospital epidemiology february 2014, vol. 35, no. 2 original article Chlorhexidine Bathing in a Tertiary Care Neonatal Intensive Care Unit: Impact on Central Line–Associated Bloodstream Infections Caroline Quach, MD, MSc;1,2,3 Aaron M. -
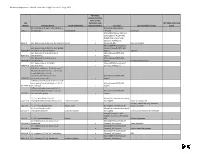
Chemicals of High Concern List (Sorted Alphabetically), July 2010
Minnesota Department of Health, Chemicals of High Concern list, July 1, 2010 Persistent, Bioaccumulative, Toxic or very CAS Persistent, very HPV (2006 and 3 of 4 Number Chemical Name Health endpoint(s) Bioaccumulative Source(s) Use example(s) or class years) (S)-4-hydroxy-3-(3-oxo-1-phenylbutyl)-2- Maine (EU Reproductive 5543-57-7 benzopyrone Reproduction Toxicant) Sunscreen Maine (CA Prop 65; IARC; EU Carcinogen; NTP 11th ROC; OSPAR Chemicals of High Concern); WA Appen1; 91-94-1 [1,1'-biphenyl]-4,4'-diamine, 3,3'-dichloro-Cancer x Minnesota HRL Dye, curing agent Maine (OSPAR Chemicals of [1,1'-biphenyl]-4,4'-diamine, N,N'-bis(2,4- Concern; Canada PBiT); WA 29398-96-7 dinitrophenyl)-3,3'-dimethoxy- x Appen1 Colorant [1,1'-Biphenyl]-4-ol, 3,4',5-tris(1,1- Maine (Canada PBiT); WA 6257-39-2 dimethylethyl)- x Appen1 [1,1'-Biphenyl]-4-ol, 3,4'-bis(1,1- Maine (Canada PBiT); WA 42479-88-9 dimethylethyl)- x Appen1 Chemical intermediate [1,1'-biphenyl]-4-ol, 3,5-bis(1,1- Maine (OSPAR Chemicals of 2668-47-5 dimethylethyl)- x Concern); WA Appen1 [2,6'-Bibenzothiazole]-7-sulfonic acid, 2'- (4-aminophenyl)-6-methyl-, diazotized, coupled with diazotized 4- aminobenzenesulfonic acid and Maine (Canada PBiT); WA 91696-90-1 resorcinol, sodium salts x Appen1 1(2H)-Quinolineethanol, 6-[(2-chloro-4,6- dinitrophenyl) azo]-3,4-dihydro-2,2,4,7- Maine (Canada PBiT); WA 63133-84-6 tetramethyl- x Appen1 1(2H)-Quinolinepropanamide, 6-(2,2- dicyanoethenyl)-3, 4-dihydro-2,2,4,7- Maine (Canada PBiT); WA 63467-15-2 tetramethyl-N-phenyl- x Appen1 1,1,1,2-Tetrachloro-2,2-bis(4- -

Guideline for Disinfection and Sterilization in Healthcare Facilities, 2008
Guideline for Disinfection and Sterilization in Healthcare Facilities, 2008 Guideline for Disinfection and Sterilization in Healthcare Facilities, 2008 William A. Rutala, Ph.D., M.P.H.1,2, David J. Weber, M.D., M.P.H.1,2, and the Healthcare Infection Control Practices Advisory Committee (HICPAC)3 1Hospital Epidemiology University of North Carolina Health Care System Chapel Hill, NC 27514 2Division of Infectious Diseases University of North Carolina School of Medicine Chapel Hill, NC 27599-7030 1 Guideline for Disinfection and Sterilization in Healthcare Facilities, 2008 3HICPAC Members Robert A. Weinstein, MD (Chair) Cook County Hospital Chicago, IL Jane D. Siegel, MD (Co-Chair) University of Texas Southwestern Medical Center Dallas, TX Michele L. Pearson, MD (Executive Secretary) Centers for Disease Control and Prevention Atlanta, GA Raymond Y.W. Chinn, MD Sharp Memorial Hospital San Diego, CA Alfred DeMaria, Jr, MD Massachusetts Department of Public Health Jamaica Plain, MA James T. Lee, MD, PhD University of Minnesota Minneapolis, MN William A. Rutala, PhD, MPH University of North Carolina Health Care System Chapel Hill, NC William E. Scheckler, MD University of Wisconsin Madison, WI Beth H. Stover, RN Kosair Children’s Hospital Louisville, KY Marjorie A. Underwood, RN, BSN CIC Mt. Diablo Medical Center Concord, CA This guideline discusses use of products by healthcare personnel in healthcare settings such as hospitals, ambulatory care and home care; the recommendations are not intended for consumer use of the products discussed. 2 -
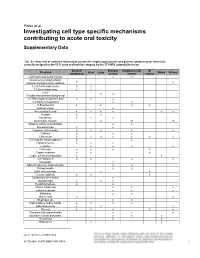
Investigating Cell Type Specific Mechanisms Contributing to Acute Oral Toxicity
Prieto et al.: Investigating cell type specific mechanisms contributing to acute oral toxicity Supplementary Data1 Tab. S1: Overview of collected information on specific target organ/system and general cytotoxicity for chemicals correctly assigned to the CLP acute oral toxicity category by the 3T3 NRU cytotoxicity assay General Nervous Cardiovascular GI Chemical Liver Lung Blood Kidney cytotoxicity system system system (±)-Propranolol hydrochloride x ax (4-ammonio-m-tolyl)ethyl(2- x x hydroxyethyl)ammonium sulphate 1,2,4-Trichlorobenzene x x 1,2-Dichlorobenzene x x 2,4,6- x x Tris(dimethylaminomethyl)phenol 2,4-Dichlorophenoxyacetic acid x x x 5,5-Diphenylhydantoin x x 5-Fluorouracil x x x x Acetophenone x Acetylsalicylic acid x x x x x x Acrolein x x Acrylamide x x x Ammonium chloride x ax ax Atropine sulfate monohydrate x x Benzaldehyde x x Cadmium (III) chloride x x x x x Caffeine x x x Chloroform x x ax x x x x Chloroquine bis(phosphate) x x Chlorpromazine x x x Codeine x x x x Colchicine x x x x Copper sulphate x x x Cupric sulfate pentahydrate x x Cyclosporin A x x x x Diazepam x Diphenhydramine hydrochloride x x Disopyramide x x Ethyl chloroacetate x x Ferrous sulphate x x x Glufosinate-ammonium x Glutethimide x ax x Hexachlorophene x x Lithium Carbonate x x x Lithium sulphate x x x Malathion x x Maleic acid x x Meprobamate x x Orphenadrine hydrochloride x x x x p-Benzoquinone x x x Phenol x x x x x Procainamide hydrochloride x x x Quinidine sulfate dehydrate x x x Resorcinol x x Rifampicin x x doi:10.14573/altex.1805181s2 ALTEX ##(#), SUPPLEMENTARY DATA 1 General Nervous Cardiovascular GI Chemical Liver Lung Blood Kidney cytotoxicity system system system Sodium Cyanate x x sodium oxalate x x x Sodium valproate x bx x x Thioridazine hydrochloride x x Valproic acid x x x x x GI: Gastrointestinal; CLP: Classification, labelling and packaging; NRU: Neutral Red Uptake; a Indirect effect: b chronic effect Tab. -

2019 Minnesota Chemicals of High Concern List
Minnesota Department of Health, Chemicals of High Concern List, 2019 Persistent, Bioaccumulative, Toxic (PBT) or very Persistent, very High Production CAS Bioaccumulative Use Example(s) and/or Volume (HPV) Number Chemical Name Health Endpoint(s) (vPvB) Source(s) Chemical Class Chemical1 Maine (CA Prop 65; IARC; IRIS; NTP Wood and textiles finishes, Cancer, Respiratory 11th ROC); WA Appen1; WA CHCC; disinfection, tissue 50-00-0 Formaldehyde x system, Eye irritant Minnesota HRV; Minnesota RAA preservative Gastrointestinal Minnesota HRL Contaminant 50-00-0 Formaldehyde (in water) system EU Category 1 Endocrine disruptor pesticide 50-29-3 DDT, technical, p,p'DDT Endocrine system Maine (CA Prop 65; IARC; IRIS; NTP PAH (chem-class) 11th ROC; OSPAR Chemicals of Concern; EuC Endocrine Disruptor Cancer, Endocrine Priority List; EPA Final PBT Rule for 50-32-8 Benzo(a)pyrene x x system TRI; EPA Priority PBT); Oregon P3 List; WA Appen1; Minnesota HRV WA Appen1; Minnesota HRL Dyes and diaminophenol mfg, wood preservation, 51-28-5 2,4-Dinitrophenol Eyes pesticide, pharmaceutical Maine (CA Prop 65; IARC; NTP 11th Preparation of amino resins, 51-79-6 Urethane (Ethyl carbamate) Cancer, Development ROC); WA Appen1 solubilizer, chemical intermediate Maine (CA Prop 65; IARC; IRIS; NTP Research; PAH (chem-class) 11th ROC; EPA Final PBT Rule for 53-70-3 Dibenzo(a,h)anthracene Cancer x TRI; WA PBT List; OSPAR Chemicals of Concern); WA Appen1; Oregon P3 List Maine (CA Prop 65; NTP 11th ROC); Research 53-96-3 2-Acetylaminofluorene Cancer WA Appen1 Maine (CA Prop 65; IARC; IRIS; NTP Lubricant, antioxidant, 55-18-5 N-Nitrosodiethylamine Cancer 11th ROC); WA Appen1 plastics stabilizer Maine (CA Prop 65; IRIS; NTP 11th Pesticide (EPA reg. -

June 16, 2014 Colleen Rogers Center for Drug Evaluation and Research
June 16, 2014 Colleen Rogers Center for Drug Evaluation and Research Food and Drug Administration Building 22, Room 5411 10903 New Hampshire Avenue Silver Spring, MD 20993 Re: Proposed Rule: Proposed Amendment of the Tentative Final Monograph, Federal Register, Vol. 78, No. 242, Tuesday, December 17, 2013. Docket identification (ID) number: FDA-1975-N-0012 Regulatory Information Number: 0910-AF69 Dear Ms. Rogers: The American Cleaning Institute (ACI)1 appreciates this opportunity to provide comments on the proposed rule to amend the 1994 tentative final monograph (the 1994 TFM) for over-the-counter (OTC) antiseptic drug products to establish conditions under which OTC consumer antiseptic products intended for use with water (referred to throughout as consumer antiseptic washes) are generally recognized as safe and effective. ACI has a specific interest in triclosan (TCS) within the proposed rule since our members produce consumer antiseptic wash products containing triclosan and manufacture triclosan. Triclosan-containing consumer antiseptic wash products play a beneficial role in the daily hygiene routines of millions of people throughout the U.S. and worldwide. They have been and are used safely and effectively in homes, hospitals, schools and workplaces every single day. Furthermore, triclosan and products containing it are regulated by a number of governmental bodies around the world and have a long track record of human and environmental safety which is supported by a multitude of science-based, transparent risk analyses. ACI members are concerned that FDA has not appropriately assessed the safety data that are available prior to proposing that additional safety data are necessary to support the safety of triclosan for this use. -

General Guidelines for NOI Forms B Through L and CWB NOI General Form (CWBNOI General.Pdf)
State of Hawaii Do NOT submit Department of Health Clean Water Branch this document. General Guidelines for NOI Forms B through L and CWB NOI General Form (CWBNOI_General.pdf) General Guidelines for Notice of Intent for Hawaii Administrative Rules, Chapter 11-55, Appendices B through L National Pollutant Discharge Elimination System (NPDES) Notice of General Permit Coverage (NGPC) For coverage under a specific NPDES General Permit, the following items are required to be submitted to the Clean Water Branch (CWB): A. CWB NOI General Form (CWBNOI_General.pdf) with Certifying Person’s original signature [via “Submit via Email” button and hard copy] B. General Permit Specific CWB NOI Form B, C, D, E, F, G, H, I, K, or L (CWBNOI_B.pdf through CWBNOI_L.doc) [via “Submit via Email” button, as applicable, and hard copy] C. All applicable attachments [via hard copy] D. $500 Filing Fee [Check made payable to “State of Hawaii”] E. Additional copies as required for Islands other than Oahu [see Notes V.D. and V.E. of the General Guidelines] TABLE OF CONTENTS Note Page General Information Applicable to All NOI Forms ......................................... 3 I. Introduction to the NPDES General Permit ........................................... 3 II. Class of Receiving State Waters Not Covered by NPDES General Permits .................. 4 III. Discharge Activities Covered by an NPDES General Permit .............................. 4 A. HAR, Chapter 11-55, Appendix B .............................................. 4 B. HAR, Chapter 11-55, Appendix C .............................................. 6 C. HAR, Chapter 11-55, Appendix D .............................................. 7 D. HAR, Chapter 11-55, Appendix E .............................................. 7 E. HAR, Chapter 11-55, Appendix F .............................................. 7 F. HAR, Chapter 11-55, Appendix G ............................................ -

WHO Model Prescribing Information Drugs Used in Skin Diseases
WHO Model Prescribing Information Drugs used in Skin Diseases World Health Organization Geneva 1997 Contents Preface 1 Introduction 3 Parasitic infections 5 Pediculosis 5 Scabies 6 Cutaneous larva migrans (creeping eruption) 7 Gnathostomiasis 8 Insect and arachnid bites and stings 9 Mosquitos and other biting flies 9 Bees, wasps, hornets and ants 10 Bedbugs and reduviid bugs 11 Scorpions 11 Poisonous spiders 12 Chiggers or harvest mites 12 Ticks 13 Superficial fungal infections 14 Dermatophyte infections 14 Pityriasis (tinea) versicolor 16 Candidosis 17 Subcutaneous fungal infections 20 Sporotrichosis 20 Mycetoma 20 Chromomycosis 21 Subcutaneous zygomycosis 21 Bacterial infections 23 Staphylococcal and streptococcal infections 23 Yaws and pinta 25 Viral infections 27 Warts 27 Herpes simplex 28 Zoster and varicella 28 Molluscum contagiosum 29 Eczematous diseases 30 Contact dermatitis 30 Atopic dermatitis 31 Seborrhoeic dermatitis 32 Scaling diseases 34 Ichthyosis 34 Xerosis 34 Papulosquamous diseases 36 Lichen planus 36 Contents (continued) Pityriasis rosea 36 Psoriasis 37 Cutaneous reactions to drugs 40 Pigmentary disorders 42 Vitiligo 42 Melasma 42 Albinism 43 Premalignant lesions and malignant tumours 44 Actinic keratosis 44 Basal cell and squamous cell carcinomas 45 Malignant melanoma 45 Photodermatoses 47 Solar urticaria 47 Polymorphous light eruptions 47 Actinic prurigo 48 Chemical photodermatoses 48 Bullous dermatoses 50 Pemphigus 50 Bullous pemphigoid 51 Dermatitis herpetiformis 52 Alopecia areata 53 Urticaria 54 Conditions common