Efficient Delivery of Nuclease Proteins for Genome Editing in Human Stem
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Rnai in Primary Cells and Difficult-To-Transfect Cell Lines
Automated High Throughput Nucleofection® RNAi in Primary Cells and Difficult-to-Transfect Cell Lines Claudia Merz, Bayer Schering Pharma AG, Berlin, Germany; Andreas Schroers, amaxa AG, Cologne, Germany; Eric Willimann, Tecan AG, Männedorf, Switzerland. Introduction Materials & Methods - Workflow Using primary cells for RNAi based applications such as target identification or – validation, requires a highly efficient transfection displaying the essential steps of the automated Nucleofector® Process: technology in combination with a reliable and robust automation system. To accomplish these requirements we integrated the amaxa 1. Transfer of the cells to the Nucleocuvette™ plate, 96-well Shuttle® in a Tecan Freedom EVO® cell transfection workstation which is based on Tecan’s Freedom EVO® liquid handling 2. Addition of the siRNA, (Steps 1 and 2 could be exchanged), platform and include all the necessary components and features for unattended cell transfection. 3. Nucleofection® process, 4. Addition of medium, Count Cells 5. Transfer of transfected cells to cell culture plate for incubation ® Nucleofector Technology prior to analysis. Remove Medium The 96-well Shuttle® combines high-throughput compatibility with the Nucleofector® Technology, which is a non-viral transfection method ideally suited for primary cells and hard-to-transfect cell lines based on a combination of buffers and electrical parameters. Nucleocuvette Plate Add Nucleofector +– The basic principle and benefits of the (empty) Solution Cell of interest Gene of interest Nucleofector® -

CRISPR/Cas9-Mediated Trp53 and Brca2 Knockout
Published OnlineFirst August 16, 2016; DOI: 10.1158/0008-5472.CAN-16-1272 Cancer Tumor and Stem Cell Biology Research CRISPR/Cas9-Mediated Trp53 and Brca2 Knockout to Generate Improved Murine Models of Ovarian High-Grade Serous Carcinoma Josephine Walton1,2, Julianna Blagih3, Darren Ennis1, Elaine Leung1, Suzanne Dowson1, Malcolm Farquharson1, Laura A. Tookman4, Clare Orange5, Dimitris Athineos3, Susan Mason3, David Stevenson3, Karen Blyth3, Douglas Strathdee3, Frances R. Balkwill2, Karen Vousden3, Michelle Lockley4, and Iain A. McNeish1,4 Abstract – – There is a need for transplantable murine models of ovarian ating novel ID8 derivatives that harbored single (Trp53 / )or – – – – high-grade serous carcinoma (HGSC) with regard to mutations in double (Trp53 / ;Brca2 / ) suppressor gene deletions. In these the human disease to assist investigations of the relationships mutants, loss of p53 alone was sufficient to increase the growth between tumor genotype, chemotherapy response, and immune rate of orthotopic tumors with significant effects observed on the microenvironment. In addressing this need, we performed whole- immune microenvironment. Specifically, p53 loss increased exome sequencing of ID8, the most widely used transplantable expression of the myeloid attractant CCL2 and promoted the model of ovarian cancer, covering 194,000 exomes at a mean infiltration of immunosuppressive myeloid cell populations into – – – – depth of 400Â with 90% exons sequenced >50Â. We found no primary tumors and their ascites. In Trp53 / ;Brca2 / mutant functional mutations in genes characteristic of HGSC (Trp53, cells, we documented a relative increase in sensitivity to the PARP Brca1, Brca2, Nf1, and Rb1), and p53 remained transcriptionally inhibitor rucaparib and slower orthotopic tumor growth – – active. Homologous recombination in ID8 remained intact in compared with Trp53 / cells, with an appearance of intratumoral þ functional assays. -
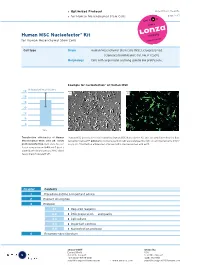
Optimized Protocol for Human Mesenchymal Stem Cells
› Optimized Protocol DPE-1001 Vs. 05-2005 › for Human Mesenchymal Stem Cells page 1 of 7 Human MSC Nucleofector® Kit for Human Mesenchymal Stem Cells Cell type Origin Human Mesenchymal Stem Cells (MSC), cryopreserved [Clonetics/BioWhittaker; Cat. No. PT-2501]. Morphology Cells with large nuclei and long spindle like protrusions.. Example for nucleofection® of Human MSC % transfection efficiency 60 A B 50 40 30 20 10 0 24 h Transfection efficiencies of Human Human MSC were nucleofected using the Human MSC Nucleofector Kit and a plasmid encoding the fluo- Mesenchymal Stem cells 24 hours rescent protein eGFP. 24 hours post-nucleofection cells were analyzed by light (A) and fluorescence micro- post nucleofection. Cells were nucleo- scopy (B). Transfection efficiencies of around 80% can be reached with eGFP. fected using program U-23 and 5 µg of a plasmid encoding the mouse MHC class I heavy chain molecule H-2Kk. Chapter Contents 1 Procedure outline & important advice 2 Product description 3 Protocol 3.1 › Required reagents 3.2 › DNA preparation and quality 3.3 › Cell culture 3.4 › Important controls 3.5 › Nucleofection protocol 4 Recommended literature 5 amaxa GmbH amaxa Inc. Europe/World USA Scientific Support Scientific Support +49 (0)221-99199-400 (240) 632-9110 [email protected] › www.amaxa.com [email protected] › Optimized Protocol DPE-1001 Vs. 05-2005 › for Human Mesenchymal Stem Cells page 2 of 7 1 Procedure outline & important advice Procedure outline Important advice 1. Preparation of cells. › Use MSCGM BulletKit (stored < 2 d at 4°C) (For details see 3.3.) › Passage interval: after reaching 70% confluency. -

NIH Public Access Author Manuscript Nat Protoc
NIH Public Access Author Manuscript Nat Protoc. Author manuscript; available in PMC 2013 June 17. NIH-PA Author ManuscriptPublished NIH-PA Author Manuscript in final edited NIH-PA Author Manuscript form as: Nat Protoc. ; 7(1): 171–192. doi:10.1038/nprot.2011.431. A Transcription Activator-Like Effector (TALE) Toolbox for Genome Engineering Neville E. Sanjana1,*, Le Cong1,2,*, Yang Zhou1,*, Margaret M. Cunniff1, Guoping Feng1, and Feng Zhang1,† 1Broad Institute of MIT and Harvard, Cambridge, MA, USAMcGovern Institute for Brain Research, Department of Brain and Cognitive Sciences, Massachusetts Institute of Technology, Cambridge, MA, USA 2Program in Biological and Biomedical Sciences, Harvard Medical School, Boston, MA, USA Abstract Transcription activator-like effectors (TALEs) are a class of naturally occurring DNA binding proteins found in the plant pathogen Xanthomonas sp. The DNA binding domain of each TALE consists of tandem 34-amino acid repeat modules that can be rearranged according to a simple cipher to target new DNA sequences. Customized TALEs can be used for a wide variety of genome engineering applications, including transcriptional modulation and genome editing. Here we describe a toolbox for rapid construction of custom TALE transcription factors (TALE-TFs) and nucleases (TALENs) using a hierarchical ligation procedure. This toolbox facilitates affordable and rapid construction of custom TALE-TFs and TALENs within one week and can be easily scaled up to construct TALEs for multiple targets in parallel. We also provide details for testing the activity in mammalian cells of custom TALE-TFs and TALENs using, respectively, qRT-PCR and Surveyor nuclease. The TALE toolbox described here will enable a broad range of biological applications. -

Methods of Transfection with Messenger RNA Gene Vectors
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Spiral - Imperial College Digital Repository Chapter 2 Methods of Transfection with Messenger RNA Gene Vectors Oleg E. Tolmachov and Tanya Tolmachova Additional information is available at the end of the chapter http://dx.doi.org/10.5772/61688 Abstract Non-viral gene delivery vectors with messenger RNA (mRNA) as a carrier of genetic information are among the staple gene transfer vectors for research in gene therapy, gene vaccination and cell fate reprogramming. As no passage of genetic cargo in and out of the nucleus is required, mRNA-based vectors typically offer the following five advantages: 1) fast start of transgene expression; 2) ability to express genes in non- dividing cells with an intact nuclear envelope; 3) insensitivity to the major gene silencing mechanisms, which operate in the nucleus; 4) absence of potentially mutagenic genomic insertions; 5) high cell survival rate after transfection procedures, which do not need to disturb nuclear envelope. In addition, mRNA-based vectors offer a simple combination of various transgenes through mixing of several mRNAs in a single multi-gene cocktail or expression of a number of proteins from a single mRNA molecule using internal ribosome entry sites (IRESes), ribosome skipping sequences and proteolytic signals. However, on the downside, uncontrolled extrac‐ ellular and intracellular decay of mRNA can be a substantial hurdle for mRNA- mediated gene transfer. Procedures for mRNA delivery are analogous to DNA transfer methods, which are well-established. In general, there are three actors in the gene delivery play, namely, the vector, the cell and the transfer environment. -

Nucleofection Protocol
NUCLEOFECTION PROTOCOL Nucleofection (electroporation) of Cas9/ synthetic RNA ribonucleoprotein (RNP) complexes for CRISPR/Cas9 genome editing (Lonza Nucleofection™ System) BACKGROUND This protocol describes how to transfect cultured cells with ribonucleoprotein (RNP) complexes that consist of purified Cas9 nuclease duplexed with synthetic guide RNA (gRNA; synthetic sgRNA or annealed crRNA and tracrRNA) using the Lonza 4D Nucleofector™ unit with 16-well Nucleocuvette™ Strips. Delivery of RNPs means that CRISPR components exist only transiently inside the cell, limiting Cas9 and guide RNA expression – this allows for the highest levels of editing efficiency and greatly reduces the chances of possible off-target and toxic effects. Furthermore, the use of synthetic guide RNAs eliminates the risk of incorporating foreign DNA into the host genome, which can occur when using plasmid-based guides. Synthego synthetic guide RNAs are of the highest quality and offer a superior alternative to in vitro transcribed (IVT) guide RNAs that are of variable quality and produce inconsistent editing results. MATERIALS REQUIRED Reagent/Material Vendor Synthego CRISPRevolution synthetic guide RNA Synthego (sgRNA or crRNA/tracrRNA) Synthego 2NLS-Cas9 nuclease Synthego Lonza Nucleofector 4D Electroporation System Lonza (#AAF-1002B and variants) Lonza SF Cell Line 4D - Nucleofector™ X Kit S Lonza (#V4XC-2032) with 16-well Nucleocuvette™ Strip DMEM, high glucose, with GlutaMAX™ Thermo Fisher (#10566 and variants) Fetal Bovine Serum (FBS) Thermo Fisher (#10437010 and variants) TrypLE™ Express Enzyme Thermo Fisher (#12605010 and variants) 1X PBS (without Ca²⁺ and Mg²⁺) Common lab supplier, or make in house Sterile tissue culture plates (24-well) Common lab supplier Microcentrifuge tubes Common lab supplier 1 synthego.com | [email protected] 20161208 NUCLEOFECTION PROTOCOL IMPORTANT CONSIDERATIONS • Wearing gloves and using nuclease-free tubes and reagents is recommended in order to avoid RNase contamination. -

California State University, Northridge
CALIFORNIA STATE UNIVERSITY, NORTHRIDGE Characterization of Chromosomal Alterations Using a Zinc-Finger Nuclease Targeting the Beta-globin Gene Locus in Hematopoietic Stem/Progenitor Cells A thesis submitted in partial fulfillment of the requirements For the degree of Master of Science in Biology By Joseph D. Long May 2017 The thesis of Joseph Long is approved: Dr. Jonathan Kelber Date Dr. Virginia Vandergon Date Dr. Cindy S. Malone, Chair Date California State University, Northridge ii Acknowledgements First, I want to thank Dr. Cindy Malone for making this opportunity and ultimately this work possible. The experiences I have had and everything that I have learned from Dr. Malone and at UCLA through the Bridges program have been invaluable to becoming a better scientist and person. While the work presented here was done through the Kohn Lab at UCLA, it was based on a solid foundation of skills learned in Dr. Malone’s lab and in courses taught by Dr. Virginia Vandergon and Dr. Jonathan Kelber at CSUN. This foundation enabled me to achieve a higher level of scientific development at UCLA that I would not have been able to reach otherwise. Second, I want to thank Dr. Donald Kohn, Dr. Zulema Romero and Dr. Caroline Kuo for making this work possible and for their incredible help and advice throughout the course of this project. Also, I want to thank everyone else in the Kohn lab for all of their contributions to this project. iii Table of Contents Signature Page…………………………………………….………………………...….....ii Acknowledgments…………………………………..………...………………….....……iii -
DNA Targeting Specificity of RNA-Guided Cas9 Nucleases
DNA targeting specificity of RNA-guided Cas9 nucleases The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation Hsu, Patrick D., David A. Scott, Joshua A. Weinstein, et al. "DNA targeting specificity of RNA-guided Cas9 nucleases." Nature Biotechnology 31:9 (2013) p.827-834. As Published http://dx.doi.org/10.1038/nbt.2647 Publisher Nature Publishing Group Version Author's final manuscript Citable link http://hdl.handle.net/1721.1/102691 Terms of Use Article is made available in accordance with the publisher's policy and may be subject to US copyright law. Please refer to the publisher's site for terms of use. HHS Public Access Author manuscript Author Manuscript Author ManuscriptNat Biotechnol Author Manuscript. Author manuscript; Author Manuscript available in PMC 2014 March 30. Published in final edited form as: Nat Biotechnol. 2013 September ; 31(9): 827–832. doi:10.1038/nbt.2647. DNA targeting specificity of RNA-guided Cas9 nucleases Patrick D Hsu1,2,3,9, David A Scott1,2,9, Joshua A Weinstein1,2, F Ann Ran1,2,3, Silvana Konermann1,2, Vineeta Agarwala1,4,5, Yinqing Li1,2, Eli J Fine6, Xuebing Wu7, Ophir Shalem1,2, Thomas J Cradick6, Luciano A Marraffini8, Gang Bao6, and Feng Zhang1,2 1Broad Institute of MIT and Harvard, Cambridge, Massachusetts, USA 2McGovern Institute for Brain Research, Department of Brain and Cognitive Sciences, Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA 3Department of Molecular -

Physical Non-Viral Gene Delivery Methods for Tissue Engineering
HHS Public Access Author manuscript Author ManuscriptAuthor Manuscript Author Ann Biomed Manuscript Author Eng. Author Manuscript Author manuscript; available in PMC 2016 November 09. Published in final edited form as: Ann Biomed Eng. 2013 March ; 41(3): 446–468. doi:10.1007/s10439-012-0678-1. Physical non-viral gene delivery methods for tissue engineering Adam J. Mellott, B.S.1, M. Laird Forrest, Ph.D.2, and Michael S. Detamore, Ph.D.1,3 1Bioengineering Program, University of Kansas, Lawrence, Kansas 66045 2Department of Pharmaceutical Chemistry, University of Kansas, Lawrence, Kansas 66047 3Department of Chemical and Petroleum Engineering, University of Kansas, Lawrence, Kansas 66045 Abstract The integration of gene therapy into tissue engineering to control differentiation and direct tissue formation is not a new concept; however, successful delivery of nucleic acids into primary cells, progenitor cells, and stem cells has proven exceptionally challenging. Viral vectors are generally highly effective at delivering nucleic acids to a variety of cell populations, both dividing and non- dividing, yet these viral vectors are marred by significant safety concerns. Non-viral vectors are preferred for gene therapy, despite lower transfection efficiencies, and possess many customizable attributes that are desirable for tissue engineering applications. However, there is no single non- viral gene delivery strategy that “fits-all” cell types and tissues. Thus, there is a compelling opportunity to examine different non-viral vectors, especially physical vectors, and compare their relative degrees of success. This review examines the advantages and disadvantages of physical non-viral methods (i.e., microinjection, ballistic gene delivery, electroporation, sonoporation, laser irradiation, magnetofection, and electric field-induced molecular vibration), with particular attention given to electroporation because of its versatility, with further special emphasis on Nucleofection™. -

Designing an Rnai Experiment Using Nucleofection™ Technical Reference Guide
BioResearch Designing an RNAi Experiment Using Nucleofection™ Technical Reference Guide The Nucleofector™ Technology is well suited for the transfection of siRNA duplexes or shRNA vectors into both primary cells and difficult-to-transfect cell lines. Choose siRNA Choose Cell Type and Transfection Protocol – Select gene target(s) – Select cell type(s) to maximize physiological relevance – Select control siRNA of results – Negative control siRNA – – Find Optimized Nucleofection™ Protocols at e.g., Thermo Scientific siGENOME Non-Targeting Control www.lonza.com/cell-database – Positive control siRNA – – Select transfection controls e.g., siGENOME® GAPDH siRNA – Untreated sample (no siRNA and transfection) – Mock-transfection (no siRNA, only transfection) ➔ ➔ Confirm siRNA Delivery Choose Detection Assay – Confirm siRNA delivery efficiency using: – Select detection assay(s) – Fluorescently-labeled siRNA – mRNA – branched-DNA, RT-PCR or – Protein – ELISA, Western, FACS analysis – Fluorescent expression plasmid (e.g., pmaxGFP™ Vector) – Phenotype – viability, apoptosis or – pmaxGFP™ Vector and maxGFP™ Reporter Protein siRNA or siRNA targeting housekeeping gene ➔ ➔ Optimize Target Knockdown Adapt Assay Conditions – Determine optimal siRNA concentration – Optimize detection assay(s) conditions for – Nucleofector™ Device ➞ 0.2 – 200 pmol (2 nM – 2 µM) specific system ➞ – 96-well Shuttle™ Device 0.04 – 40 pmol (2 nM – 2 µM) – Determine optimal cell densities for linear detection range – Correlate results from multiple assays ➔ ➔ Optimize Assay Conditions -

An Efficient Transfection Method for Mouse Embryonic Stem Cells
Gene Therapy (2009) 16, 154–158 & 2009 Macmillan Publishers Limited All rights reserved 0969-7128/09 $32.00 www.nature.com/gt SHORT COMMUNICATION An efficient transfection method for mouse embryonic stem cells BS Ko1,2,5, TC Chang1,5, SK Shyue3, YC Chen4 and JY Liou1 1National Health Research Institutes, Zhunan Town, Miaoli County, Taiwan; 2Department of Internal Medicine, National Taiwan University Hospital, Taipei, Taiwan; 3Institutes of Biomedical Science, Academic Sinica, Taipei, Taiwan and 4Department of Laboratory Medicine, National Taiwan University Hospital, Taipei, Taiwan Embryonic stem (ES) cells are considered to have potentials Our results show that transfection by Effectene achieves the for tissue regeneration and treatment of diverse human efficiency of 498% in CCE and 480% in D3 cells. The diseases. ES cells are capable of indefinite renewal and optimal ratio of DNA:Effectene for EGFP transfection is proliferation, which can be induced to differentiate into tissues between 1:4 and 1:8. Transient-expressed EGFP or endo- of all three germ lines. Despite these exciting potential, it genous protein kinase A (PKA) were significantly knocked remains unclear as to how the renewal and differentiation down by Effectene transfection of specific siRNA. High EGFP programs are operated and regulated at the genetic level. level expression and accumulation in mES cells induces Genetic manipulation such as delivery of exogenous gene minor cytotoxicity but can be reduced by introducing siRNA of expression or knockdown with small interfering RNA (siRNA) EGFP. Further, this transfection method did not significantly is commonly used in most of cancer or transformed cells but affect mES properties of proliferation or differentiation. -

CRISPR/Cas9-Mediated Trp53 and Brca2 Knockout to Generate Improved Murine Models of Ovarian High-Grade Serous Carcinoma
Published OnlineFirst August 16, 2016; DOI: 10.1158/0008-5472.CAN-16-1272 Cancer Tumor and Stem Cell Biology Research CRISPR/Cas9-Mediated Trp53 and Brca2 Knockout to Generate Improved Murine Models of Ovarian High-Grade Serous Carcinoma Josephine Walton1,2, Julianna Blagih3, Darren Ennis1, Elaine Leung1, Suzanne Dowson1, Malcolm Farquharson1, Laura A. Tookman4, Clare Orange5, Dimitris Athineos3, Susan Mason3, David Stevenson3, Karen Blyth3, Douglas Strathdee3, Frances R. Balkwill2, Karen Vousden3, Michelle Lockley4, and Iain A. McNeish1,4 Abstract – – There is a need for transplantable murine models of ovarian ating novel ID8 derivatives that harbored single (Trp53 / )or – – – – high-grade serous carcinoma (HGSC) with regard to mutations in double (Trp53 / ;Brca2 / ) suppressor gene deletions. In these the human disease to assist investigations of the relationships mutants, loss of p53 alone was sufficient to increase the growth between tumor genotype, chemotherapy response, and immune rate of orthotopic tumors with significant effects observed on the microenvironment. In addressing this need, we performed whole- immune microenvironment. Specifically, p53 loss increased exome sequencing of ID8, the most widely used transplantable expression of the myeloid attractant CCL2 and promoted the model of ovarian cancer, covering 194,000 exomes at a mean infiltration of immunosuppressive myeloid cell populations into – – – – depth of 400Â with 90% exons sequenced >50Â. We found no primary tumors and their ascites. In Trp53 / ;Brca2 / mutant functional mutations in genes characteristic of HGSC (Trp53, cells, we documented a relative increase in sensitivity to the PARP Brca1, Brca2, Nf1, and Rb1), and p53 remained transcriptionally inhibitor rucaparib and slower orthotopic tumor growth – – active. Homologous recombination in ID8 remained intact in compared with Trp53 / cells, with an appearance of intratumoral þ functional assays.