Peptide Nucleic Acids As a Tool for Site-Specific Gene Editing
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Rnai in Primary Cells and Difficult-To-Transfect Cell Lines
Automated High Throughput Nucleofection® RNAi in Primary Cells and Difficult-to-Transfect Cell Lines Claudia Merz, Bayer Schering Pharma AG, Berlin, Germany; Andreas Schroers, amaxa AG, Cologne, Germany; Eric Willimann, Tecan AG, Männedorf, Switzerland. Introduction Materials & Methods - Workflow Using primary cells for RNAi based applications such as target identification or – validation, requires a highly efficient transfection displaying the essential steps of the automated Nucleofector® Process: technology in combination with a reliable and robust automation system. To accomplish these requirements we integrated the amaxa 1. Transfer of the cells to the Nucleocuvette™ plate, 96-well Shuttle® in a Tecan Freedom EVO® cell transfection workstation which is based on Tecan’s Freedom EVO® liquid handling 2. Addition of the siRNA, (Steps 1 and 2 could be exchanged), platform and include all the necessary components and features for unattended cell transfection. 3. Nucleofection® process, 4. Addition of medium, Count Cells 5. Transfer of transfected cells to cell culture plate for incubation ® Nucleofector Technology prior to analysis. Remove Medium The 96-well Shuttle® combines high-throughput compatibility with the Nucleofector® Technology, which is a non-viral transfection method ideally suited for primary cells and hard-to-transfect cell lines based on a combination of buffers and electrical parameters. Nucleocuvette Plate Add Nucleofector +– The basic principle and benefits of the (empty) Solution Cell of interest Gene of interest Nucleofector® -
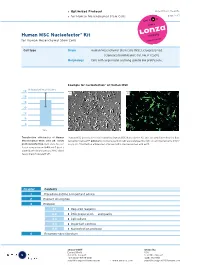
Optimized Protocol for Human Mesenchymal Stem Cells
› Optimized Protocol DPE-1001 Vs. 05-2005 › for Human Mesenchymal Stem Cells page 1 of 7 Human MSC Nucleofector® Kit for Human Mesenchymal Stem Cells Cell type Origin Human Mesenchymal Stem Cells (MSC), cryopreserved [Clonetics/BioWhittaker; Cat. No. PT-2501]. Morphology Cells with large nuclei and long spindle like protrusions.. Example for nucleofection® of Human MSC % transfection efficiency 60 A B 50 40 30 20 10 0 24 h Transfection efficiencies of Human Human MSC were nucleofected using the Human MSC Nucleofector Kit and a plasmid encoding the fluo- Mesenchymal Stem cells 24 hours rescent protein eGFP. 24 hours post-nucleofection cells were analyzed by light (A) and fluorescence micro- post nucleofection. Cells were nucleo- scopy (B). Transfection efficiencies of around 80% can be reached with eGFP. fected using program U-23 and 5 µg of a plasmid encoding the mouse MHC class I heavy chain molecule H-2Kk. Chapter Contents 1 Procedure outline & important advice 2 Product description 3 Protocol 3.1 › Required reagents 3.2 › DNA preparation and quality 3.3 › Cell culture 3.4 › Important controls 3.5 › Nucleofection protocol 4 Recommended literature 5 amaxa GmbH amaxa Inc. Europe/World USA Scientific Support Scientific Support +49 (0)221-99199-400 (240) 632-9110 [email protected] › www.amaxa.com [email protected] › Optimized Protocol DPE-1001 Vs. 05-2005 › for Human Mesenchymal Stem Cells page 2 of 7 1 Procedure outline & important advice Procedure outline Important advice 1. Preparation of cells. › Use MSCGM BulletKit (stored < 2 d at 4°C) (For details see 3.3.) › Passage interval: after reaching 70% confluency. -

Methods of Transfection with Messenger RNA Gene Vectors
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Spiral - Imperial College Digital Repository Chapter 2 Methods of Transfection with Messenger RNA Gene Vectors Oleg E. Tolmachov and Tanya Tolmachova Additional information is available at the end of the chapter http://dx.doi.org/10.5772/61688 Abstract Non-viral gene delivery vectors with messenger RNA (mRNA) as a carrier of genetic information are among the staple gene transfer vectors for research in gene therapy, gene vaccination and cell fate reprogramming. As no passage of genetic cargo in and out of the nucleus is required, mRNA-based vectors typically offer the following five advantages: 1) fast start of transgene expression; 2) ability to express genes in non- dividing cells with an intact nuclear envelope; 3) insensitivity to the major gene silencing mechanisms, which operate in the nucleus; 4) absence of potentially mutagenic genomic insertions; 5) high cell survival rate after transfection procedures, which do not need to disturb nuclear envelope. In addition, mRNA-based vectors offer a simple combination of various transgenes through mixing of several mRNAs in a single multi-gene cocktail or expression of a number of proteins from a single mRNA molecule using internal ribosome entry sites (IRESes), ribosome skipping sequences and proteolytic signals. However, on the downside, uncontrolled extrac‐ ellular and intracellular decay of mRNA can be a substantial hurdle for mRNA- mediated gene transfer. Procedures for mRNA delivery are analogous to DNA transfer methods, which are well-established. In general, there are three actors in the gene delivery play, namely, the vector, the cell and the transfer environment. -

Nucleofection Protocol
NUCLEOFECTION PROTOCOL Nucleofection (electroporation) of Cas9/ synthetic RNA ribonucleoprotein (RNP) complexes for CRISPR/Cas9 genome editing (Lonza Nucleofection™ System) BACKGROUND This protocol describes how to transfect cultured cells with ribonucleoprotein (RNP) complexes that consist of purified Cas9 nuclease duplexed with synthetic guide RNA (gRNA; synthetic sgRNA or annealed crRNA and tracrRNA) using the Lonza 4D Nucleofector™ unit with 16-well Nucleocuvette™ Strips. Delivery of RNPs means that CRISPR components exist only transiently inside the cell, limiting Cas9 and guide RNA expression – this allows for the highest levels of editing efficiency and greatly reduces the chances of possible off-target and toxic effects. Furthermore, the use of synthetic guide RNAs eliminates the risk of incorporating foreign DNA into the host genome, which can occur when using plasmid-based guides. Synthego synthetic guide RNAs are of the highest quality and offer a superior alternative to in vitro transcribed (IVT) guide RNAs that are of variable quality and produce inconsistent editing results. MATERIALS REQUIRED Reagent/Material Vendor Synthego CRISPRevolution synthetic guide RNA Synthego (sgRNA or crRNA/tracrRNA) Synthego 2NLS-Cas9 nuclease Synthego Lonza Nucleofector 4D Electroporation System Lonza (#AAF-1002B and variants) Lonza SF Cell Line 4D - Nucleofector™ X Kit S Lonza (#V4XC-2032) with 16-well Nucleocuvette™ Strip DMEM, high glucose, with GlutaMAX™ Thermo Fisher (#10566 and variants) Fetal Bovine Serum (FBS) Thermo Fisher (#10437010 and variants) TrypLE™ Express Enzyme Thermo Fisher (#12605010 and variants) 1X PBS (without Ca²⁺ and Mg²⁺) Common lab supplier, or make in house Sterile tissue culture plates (24-well) Common lab supplier Microcentrifuge tubes Common lab supplier 1 synthego.com | [email protected] 20161208 NUCLEOFECTION PROTOCOL IMPORTANT CONSIDERATIONS • Wearing gloves and using nuclease-free tubes and reagents is recommended in order to avoid RNase contamination. -

Physical Non-Viral Gene Delivery Methods for Tissue Engineering
HHS Public Access Author manuscript Author ManuscriptAuthor Manuscript Author Ann Biomed Manuscript Author Eng. Author Manuscript Author manuscript; available in PMC 2016 November 09. Published in final edited form as: Ann Biomed Eng. 2013 March ; 41(3): 446–468. doi:10.1007/s10439-012-0678-1. Physical non-viral gene delivery methods for tissue engineering Adam J. Mellott, B.S.1, M. Laird Forrest, Ph.D.2, and Michael S. Detamore, Ph.D.1,3 1Bioengineering Program, University of Kansas, Lawrence, Kansas 66045 2Department of Pharmaceutical Chemistry, University of Kansas, Lawrence, Kansas 66047 3Department of Chemical and Petroleum Engineering, University of Kansas, Lawrence, Kansas 66045 Abstract The integration of gene therapy into tissue engineering to control differentiation and direct tissue formation is not a new concept; however, successful delivery of nucleic acids into primary cells, progenitor cells, and stem cells has proven exceptionally challenging. Viral vectors are generally highly effective at delivering nucleic acids to a variety of cell populations, both dividing and non- dividing, yet these viral vectors are marred by significant safety concerns. Non-viral vectors are preferred for gene therapy, despite lower transfection efficiencies, and possess many customizable attributes that are desirable for tissue engineering applications. However, there is no single non- viral gene delivery strategy that “fits-all” cell types and tissues. Thus, there is a compelling opportunity to examine different non-viral vectors, especially physical vectors, and compare their relative degrees of success. This review examines the advantages and disadvantages of physical non-viral methods (i.e., microinjection, ballistic gene delivery, electroporation, sonoporation, laser irradiation, magnetofection, and electric field-induced molecular vibration), with particular attention given to electroporation because of its versatility, with further special emphasis on Nucleofection™. -

Designing an Rnai Experiment Using Nucleofection™ Technical Reference Guide
BioResearch Designing an RNAi Experiment Using Nucleofection™ Technical Reference Guide The Nucleofector™ Technology is well suited for the transfection of siRNA duplexes or shRNA vectors into both primary cells and difficult-to-transfect cell lines. Choose siRNA Choose Cell Type and Transfection Protocol – Select gene target(s) – Select cell type(s) to maximize physiological relevance – Select control siRNA of results – Negative control siRNA – – Find Optimized Nucleofection™ Protocols at e.g., Thermo Scientific siGENOME Non-Targeting Control www.lonza.com/cell-database – Positive control siRNA – – Select transfection controls e.g., siGENOME® GAPDH siRNA – Untreated sample (no siRNA and transfection) – Mock-transfection (no siRNA, only transfection) ➔ ➔ Confirm siRNA Delivery Choose Detection Assay – Confirm siRNA delivery efficiency using: – Select detection assay(s) – Fluorescently-labeled siRNA – mRNA – branched-DNA, RT-PCR or – Protein – ELISA, Western, FACS analysis – Fluorescent expression plasmid (e.g., pmaxGFP™ Vector) – Phenotype – viability, apoptosis or – pmaxGFP™ Vector and maxGFP™ Reporter Protein siRNA or siRNA targeting housekeeping gene ➔ ➔ Optimize Target Knockdown Adapt Assay Conditions – Determine optimal siRNA concentration – Optimize detection assay(s) conditions for – Nucleofector™ Device ➞ 0.2 – 200 pmol (2 nM – 2 µM) specific system ➞ – 96-well Shuttle™ Device 0.04 – 40 pmol (2 nM – 2 µM) – Determine optimal cell densities for linear detection range – Correlate results from multiple assays ➔ ➔ Optimize Assay Conditions -

An Efficient Transfection Method for Mouse Embryonic Stem Cells
Gene Therapy (2009) 16, 154–158 & 2009 Macmillan Publishers Limited All rights reserved 0969-7128/09 $32.00 www.nature.com/gt SHORT COMMUNICATION An efficient transfection method for mouse embryonic stem cells BS Ko1,2,5, TC Chang1,5, SK Shyue3, YC Chen4 and JY Liou1 1National Health Research Institutes, Zhunan Town, Miaoli County, Taiwan; 2Department of Internal Medicine, National Taiwan University Hospital, Taipei, Taiwan; 3Institutes of Biomedical Science, Academic Sinica, Taipei, Taiwan and 4Department of Laboratory Medicine, National Taiwan University Hospital, Taipei, Taiwan Embryonic stem (ES) cells are considered to have potentials Our results show that transfection by Effectene achieves the for tissue regeneration and treatment of diverse human efficiency of 498% in CCE and 480% in D3 cells. The diseases. ES cells are capable of indefinite renewal and optimal ratio of DNA:Effectene for EGFP transfection is proliferation, which can be induced to differentiate into tissues between 1:4 and 1:8. Transient-expressed EGFP or endo- of all three germ lines. Despite these exciting potential, it genous protein kinase A (PKA) were significantly knocked remains unclear as to how the renewal and differentiation down by Effectene transfection of specific siRNA. High EGFP programs are operated and regulated at the genetic level. level expression and accumulation in mES cells induces Genetic manipulation such as delivery of exogenous gene minor cytotoxicity but can be reduced by introducing siRNA of expression or knockdown with small interfering RNA (siRNA) EGFP. Further, this transfection method did not significantly is commonly used in most of cancer or transformed cells but affect mES properties of proliferation or differentiation. -

Interaction of Modified Oligonucleotides with Nuclear Proteins, Formation of Novel Nuclear Structures and Sequence-Independent Effects on RNA Processing
bioRxiv preprint doi: https://doi.org/10.1101/446773; this version posted October 18, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Interaction of modified oligonucleotides with nuclear proteins, formation of novel nuclear structures and sequence-independent effects on RNA processing Loren L Flynn #, Ruohan Li # May T Aung-Htut, Ianthe L Pitout, Jack Cooper, Alysia Hubbard, Lisa Griffiths, Charlie Bond, Steve D Wilton, Archa H Fox* and Sue Fletcher* # authors contributed equally *corresponding authors Abstract (max 150 words) Oligonucleotides and nucleic acid analogues that alter gene expression are showing therapeutic promise for selected human diseases. The modification of synthetic nucleic acids to protect against nuclease degradation and to influence drug function is common practice, however, such modifications may also confer unexpected physicochemical and biological properties. Here we report backbone-specific effects of modified oligonucleotides on subnuclear organelles, altered distribution of nuclear proteins, the appearance of novel structured nuclear inclusions, and modification of RNA processing in cultured cells transfected with antisense oligonucleotides on a phosphorothioate backbone. Phosphodiester and phosphorodiamidate morpholino oligomers elicited no such consequences. Disruption of subnuclear structures and proteins elicit severe phenotypic disturbances, revealed by transcriptomic analysis of fibroblasts exhibiting such disruption. These data suggest that the toxic effects and adverse events reported after clinical evaluation of phosphorothioate nucleic acid drugs may be mediated, at least in part, by non-specific interaction of nuclear components with the phosphorothioate backbone. -

Optimized Protocol for Unstimulated Human T Cells
Amaxa® Human T Cell Nucleofector® Kit For unstimulated Human T Cells Small round lymphoblastoid cells; subpopulation of human peripheral blood mononuclear cells (PBMC). PBMC should be purified from fresh human blood samples treated with an anti-coagulant or from leukocyte-enriched buffy coat Example for Nucleofection® of CD3+ T cells with different reporter genes A B C D E side scatter side CD3-APC CD3-APC PI CD3-APC forward scatter GFP FL-2 GFP GFP PBMC were freshly isolated from buffy coat and transfected with Nucleofector® Program V-024 and pmaxGFP® Vector. 24 hours post Nucleofection®, cells were analyzed by flow cytometry. Lymphocytes were gated according to forward/side scatter (A). T cells were stained with antibody directed against CD3. Dead cells were excluded by propidium iodide staining and gating (B/C). maxGFP® Protein E xpression of T cells is shown after Nucleofection® without (D) and with plasmid DNA (E). % 100 50 4 hours Transfection eciency 24 hours 48 hours 80 40 RLU/1000 cells RLU/1000 60 30 40 20 20 10 0 U-014 V-024 0 U-014 V-024 Transfection efficiency of human T cells 24 hours post Nucleofection®.Day Luciferase expression of purified human T cells. Human T cells were purified by MACS Cells were transfected with program U-014 or V-024 and 2 µg pmaxGFP® separation. Purified T cells were transfected with the Human T Cell Nucleofector® Vector. Cell viability (% PI negative) is usually 80% for both programs. Kit, program U-014 or program V-024 and 2 µg of a pGl3-CMV plasmid. -

Opportunities and Challenges in the Delivery of Mrna-Based Vaccines
pharmaceutics Review Opportunities and Challenges in the Delivery of mRNA-Based Vaccines Abishek Wadhwa , Anas Aljabbari , Abhijeet Lokras , Camilla Foged and Aneesh Thakur * Department of Pharmacy, Faculty of Health and Medical Sciences, University of Copenhagen, Universitetsparken 2, DK-2100 Copenhagen Ø, Denmark; [email protected] (A.W.); [email protected] (A.A.); [email protected] (A.L.); [email protected] (C.F.) * Correspondence: [email protected]; Tel.: + 45-3533-3938; Fax: +45-3533-6001 Received: 28 December 2019; Accepted: 26 January 2020; Published: 28 January 2020 Abstract: In the past few years, there has been increasing focus on the use of messenger RNA (mRNA) as a new therapeutic modality. Current clinical efforts encompassing mRNA-based drugs are directed toward infectious disease vaccines, cancer immunotherapies, therapeutic protein replacement therapies, and treatment of genetic diseases. However, challenges that impede the successful translation of these molecules into drugs are that (i) mRNA is a very large molecule, (ii) it is intrinsically unstable and prone to degradation by nucleases, and (iii) it activates the immune system. Although some of these challenges have been partially solved by means of chemical modification of the mRNA, intracellular delivery of mRNA still represents a major hurdle. The clinical translation of mRNA-based therapeutics requires delivery technologies that can ensure stabilization of mRNA under physiological conditions. Here, we (i) review opportunities and challenges in the delivery of mRNA-based therapeutics with a focus on non-viral delivery systems, (ii) present the clinical status of mRNA vaccines, and (iii) highlight perspectives on the future of this promising new type of medicine. -

Multifunctional Delivery Systems for Peptide Nucleic Acids
pharmaceuticals Review Multifunctional Delivery Systems for Peptide Nucleic Acids Stefano Volpi , Umberto Cancelli, Martina Neri and Roberto Corradini * Department of Chemistry, Life Sciences and Environmental Sustainability, University of Parma, 43124 Parma, Italy; [email protected] (S.V.); [email protected] (U.C.); [email protected] (M.N.) * Correspondence: [email protected]; Tel.: +39-0521-905410 Abstract: The number of applications of peptide nucleic acids (PNAs)—oligonucleotide analogs with a polyamide backbone—is continuously increasing in both in vitro and cellular systems and, parallel to this, delivery systems able to bring PNAs to their targets have been developed. This review is intended to give to the readers an overview on the available carriers for these oligonucleotide mimics, with a particular emphasis on newly developed multi-component- and multifunctional vehicles which boosted PNA research in recent years. The following approaches will be discussed: (a) conjugation with carrier molecules and peptides; (b) liposome formulations; (c) polymer nanopar- ticles; (d) inorganic porous nanoparticles; (e) carbon based nanocarriers; and (f) self-assembled and supramolecular systems. New therapeutic strategies enabled by the combination of PNA and proper delivery systems are discussed. Keywords: peptide nucleic acids; delivery; nanoparticles; conjugates; multifunctional systems 1. Introduction 1.1. Peptide Nucleic Acids and Their Uses Peptide nucleic acids (PNAs, Figure1) are DNA analogues in which the sugar- Citation: Volpi, S.; Cancelli, U.; phosphate units connecting the nucleobases are replaced by N-(2-aminoethyl)glycine Neri, M.; Corradini, R. moieties [1]. These molecules are excellent binding partners for cognate DNA and RNA Multifunctional Delivery Systems for strands, being able to exploit both the canonical Watson–Crick (WC) base pairing to form Peptide Nucleic Acids. -

Nucleofection an Efficient Non-Viral Transfection Technique Forifn-T-EGFP Gene Expression Study in Sahiwal Cattle Fibroblast Cells
Journal of Developmental Biology and Tissue Engineering Vol. 3(3), pp. 23-32, March 2011 Available online http://www.academicjournals.org/jdbte ISSN 2141-2251 ©2011 Academic Journals Full Length Research Paper Nucleofection an efficient non-viral transfection technique forIFN-t-EGFP gene expression study in Sahiwal cattle fibroblast cells Anand Laxmi N.1, Gunjan G.1 and Prateesh M.2* 1DCP, NDRI, Karnal, India. 2IVRI, Barrielly, India. Accepted 20 December, 2010 Viral based techniques were considered to be the most efficient systems to deliver DNA into fibroblast cells as they show high transgene expression in many cellular models. Viral approaches are complicated by immune response, intracellular trafficking potential mutations and genetic alterations due to integration. The nucleofector TM technology is electroporation based gene transfer technique which has been proved to be an efficient tool for transfecting primary cells and hard to transfect cell lines. The present study was designed to examine Sahiwal fibroblast cell to act as competent donor cell in gene expression studies. Using a green fluorescent protein reporter vector, a high transgene expression level was obtained using U-12 and U-23 pulsing programs: 45 and 70% respectively. Cell recoveries and viabilities were 90.5 and 95% respectively for U3-23 program. Overall transfection efficiency was 60% as observed on evaluation by flowcytometry. Further, the cells confirmed to be positive for gene expression when subjected to PCR, RT- PCR and, flow cytometry analysis using pGFP repoter other than supplied by Amaxa system. Hence, the cells were transfected with bIFN-t---GFP reporter gene construct by nucleofection technique and similarly, cells were evaluated for expression of gene by fluorescence microscopy.