Metabolic Evolution of a Deep-Branching Hyperthermophilic Chemoautotrophic Bacterium
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Diversity of Understudied Archaeal and Bacterial Populations of Yellowstone National Park: from Genes to Genomes Daniel Colman
University of New Mexico UNM Digital Repository Biology ETDs Electronic Theses and Dissertations 7-1-2015 Diversity of understudied archaeal and bacterial populations of Yellowstone National Park: from genes to genomes Daniel Colman Follow this and additional works at: https://digitalrepository.unm.edu/biol_etds Recommended Citation Colman, Daniel. "Diversity of understudied archaeal and bacterial populations of Yellowstone National Park: from genes to genomes." (2015). https://digitalrepository.unm.edu/biol_etds/18 This Dissertation is brought to you for free and open access by the Electronic Theses and Dissertations at UNM Digital Repository. It has been accepted for inclusion in Biology ETDs by an authorized administrator of UNM Digital Repository. For more information, please contact [email protected]. Daniel Robert Colman Candidate Biology Department This dissertation is approved, and it is acceptable in quality and form for publication: Approved by the Dissertation Committee: Cristina Takacs-Vesbach , Chairperson Robert Sinsabaugh Laura Crossey Diana Northup i Diversity of understudied archaeal and bacterial populations from Yellowstone National Park: from genes to genomes by Daniel Robert Colman B.S. Biology, University of New Mexico, 2009 DISSERTATION Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Biology The University of New Mexico Albuquerque, New Mexico July 2015 ii DEDICATION I would like to dedicate this dissertation to my late grandfather, Kenneth Leo Colman, associate professor of Animal Science in the Wool laboratory at Montana State University, who even very near the end of his earthly tenure, thought it pertinent to quiz my knowledge of oxidized nitrogen compounds. He was a man of great curiosity about the natural world, and to whom I owe an acknowledgement for his legacy of intellectual (and actual) wanderlust. -

FUNCTIONAL STUDIES on Csis and Csps
FUNCTIONAL STUDIES ON CSIs AND CSPs FUNCTIONAL SIGNIFICANCE OF NOVEL MOLECULAR MARKERS SPECIFIC FOR DEINOCOCCUS AND CHLAMYDIAE SPECIES BY F M NAZMUL HASSAN, B.Sc. A Thesis Submitted to the School of Graduate Studies in Partial Fulfillment of the Requirements for the Degree Master of Science McMaster University © Copyright by F M Nazmul Hassan, December, 2017 MASTER OF SCIENCE (2017) McMaster University (Biochemistry) Hamilton, Ontario TITLE: Functional Significance of Novel Molecular Markers Specific for Deinococcus and Chlamydiae Species AUTHOR: F M Nazmul Hassan, B.Sc. (Khulna University) SUPERVISOR: Professor Radhey S. Gupta NUMBER OF PAGES: xv, 108 ii This thesis is dedicated to my mom and dad. I lost my mother when I was a high school student. She always liked to share stories about science and inspired me to do something for the beneficial of human being. Her dream was guided by my father. I was always encouraged by my father to do higher education. I have lost my father this year. It was most critical moment of my life. But, I have continued to do research because of fulfill his dream. iii THESIS ABSTRACT The Deinococcus species are highly resistant to oxidation, desiccation, and radiation. Very few characteristics explain these unique features of Deinococcus species. This study reports the results of detailed comparative genomics, structural and protein-protein interactions studies on the DNA repair proteins from Deinococcus species. Comparative genomics studies have identified a large number of conserved signature indels (CSIs) in the DNA repair proteins that are specific for Deinococcus species. In parallel, I have carried out the structural and protein-protein interactions studies of CSIs which are present in nucleotide excision repair (NER), UV damage endonuclease (UvsE)-dependent excision repair (UVER) and homologous recombination (HR) pathways proteins. -

Reduction by the Methylreductase System in Methanobacterium Bryantii WILLIAM B
JOURNAL OF BACTERIOLOGY, Jan. 1987, p. 87-92 Vol. 169, No. 1 0021-9193/87/010087-06$02.00/0 Copyright © 1987, American Society for Microbiology Inhibition by Corrins of the ATP-Dependent Activation and CO2 Reduction by the Methylreductase System in Methanobacterium bryantii WILLIAM B. WHITMAN'* AND RALPH S. WOLFE2 Department of Microbiology, University of Georgia, Athens, Georgia 30602,1 and Department of Microbiology, University ofIllinois, Urbana, Illinois 618012 Received 1 August 1986/Accepted 28 September 1986 Corrins inhibited the ATP-dependent activation of the methylreductase system and the methyl coenzyme M-dependent reduction of CO2 in extracts of Methanobacterium bryantii resolved from low-molecular-weight factors. The concentrations of cobinamides and cobamides required for one-half of maximal inhibition of the ATP-depen4ent activation were between 1 and 5 ,M. Cobinamides were more inhibitory at lower concentra- tiops than cobamides. Deoxyadenosylcobalamin was not inhibitory at concentrations up to 25 ,uM. The inhibition of CO2 reduction was competitive with respect to CO2. The concentration of methylcobalamin required for one-half of maximal inhibition was 5 ,M. Other cobamideg inhibited at similar concentrations, but diaquacobinami4e inhibited at lower concentrations. With respect to their affinities and specificities for corrins, inhibition of both the ATP-dependent activation'and CO2 reduction closely resembled the corrin- dependent activation of the methylreductase described in similar extracts (W. B. Whitman and R. S. Wolfe, J. Bacteriol. 164:165-172, 1985). However, whether the multiple effects of corrins are due to action at a single site is unknown. The effect of corrins (cobamides and cobinamides) on in CO2 reduction. -

A Tertiary-Branched Tetra-Amine, N4-Aminopropylspermidine Is A
Journal of Japanese Society for Extremophiles (2010) Vol.9 (2) Journal of Japanese Society for Extremophiles (2010) Vol. 9 (2), 75-77 ORIGINAL PAPER a a b Hamana K , Hayashi H and Niitsu M NOTE 4 A tertiary-branched tetra-amine, N -aminopropylspermidine is a major cellular polyamine in an anaerobic thermophile, Caldisericum exile belonging to a new bacterial phylum, Caldiserica a Faculty of Engineering, Maebashi Institute of Technology, Maebashi, Gunma 371-0816, Japan. b Faculty of Pharmaceutical Sciences, Josai University, Sakado, Saitama 350-0290, Japan. Corresponding author: Koei Hamana, [email protected] Phone: +81-27-234-4611, Fax: +81-27-234-4611 Received: November 17, 2010 / Revised: December 8, 2010 /Accepted: December 8, 2010 Abstract Acid-extractable cellular polyamines of Anaerobic, moderately thermophilic, filamentous, thermophilic Caldisericum exile belonging to a new thiosulfate-reducing Caldisericum exile was isolated bacterial phylum, Caldiserica were analyzed by HPLC from a terrestrial hot spring in Japan for the first and GC. The coexistence of an unusual tertiary cultivated representative of the candidate phylum OP5 4 brancehed tetra-amine, N -aminopropylspermidine with and located in the newly validated bacterial phylum 19,20) spermine, a linear tetra-amine, as the major polyamines Caldiserica (order Caldisericales) . The in addition to putrescine and spermidine, is first reported temperature range for growth is 55-70°C, with the 20) in the moderate thermophile isolated from a terrestrial optimum growth at 65°C . The optimum growth 20) hot spring in Japan. Linear and branched penta-amines occurs at pH 6.5 and with the absence of NaCl . T were not detected. -
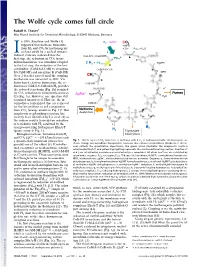
The Wolfe Cycle Comes Full Circle
The Wolfe cycle comes full circle Rudolf K. Thauer1 Max Planck Institute for Terrestrial Microbiology, D-35043 Marburg, Germany n 1988, Rouvière and Wolfe (1) H - ΔμNa+ 2 CO2 suggested that methane formation + MFR from H and CO by methanogenic + 2H+ *Fd + H O I 2 2 ox 2 archaea could be a cyclical process. j O = Indirect evidence indicated that the CoB-SH + CoM-SH fi *Fd 2- a rst step, the reduction of CO2 to for- red R mylmethanofuran, was somehow coupled + * H MPT 2 H2 Fdox 4 to the last step, the reduction of the het- h erodisulfide (CoM-S-S-CoB) to coenzyme CoM-S-S-CoB b MFR M (CoM-SH) and coenzyme B (CoB-SH). H Over 2 decades passed until the coupling C 4 10 mechanism was unraveled in 2011: Via g flavin-based electron bifurcation, the re- CoB-SH duction of CoM-S-S-CoB with H provides 2 H+ the reduced ferredoxin (Fig. 1h) required c + Purines for CO2 reduction to formylmethanofuran ΔμNa + H MPT 4 f H O (2) (Fig. 1a). However, one question still 2 remained unanswered: How are the in- termediates replenished that are removed CoM-SH for the biosynthesis of cell components H Methionine d from CO2 (orange arrows in Fig. 1)? This Acetyl-CoA e anaplerotic (replenishing) reaction has F420 F420H2 recently been identified by Lie et al. (3) as F420 F420H2 the sodium motive force-driven reduction H i of ferredoxin with H2 catalyzed by the i energy-converting hydrogenase EhaA-T H2 (green arrow in Fig. -

Complete Genome Sequence of the Thermophilic Thermus Sp
Teh et al. Standards in Genomic Sciences (2015) 10:76 DOI 10.1186/s40793-015-0053-6 SHORT GENOME REPORT Open Access Complete genome sequence of the thermophilic Thermus sp. CCB_US3_UF1 from a hot spring in Malaysia Beng Soon Teh1,5*, Nyok-Sean Lau1*, Fui Ling Ng2, Ahmad Yamin Abdul Rahman2, Xuehua Wan3, Jennifer A. Saito3, Shaobin Hou3, Aik-Hong Teh1, Nazalan Najimudin2 and Maqsudul Alam3,4ˆ Abstract Thermus sp. strain CCB_US3_UF1 is a thermophilic bacterium of the genus Thermus, a member of the family Thermaceae. Members of the genus Thermus have been widely used as a biological model for structural biology studies and to understand the mechanism of microbial adaptation under thermal environments. Here, we present the complete genome sequence of Thermus sp. CCB_US3_UF1 isolated from a hot spring in Malaysia, which is the fifth member of the genus Thermus with a completely sequenced and publicly available genome (Genbank date of release: December 2, 2011). Thermus sp. CCB_US3_UF1 has the third largest genome within the genus. The complete genome comprises of a chromosome of 2.26 Mb and a plasmid of 19.7 kb. The genome contains 2279 protein-coding and 54 RNA genes. In addition, its genome revealed potential pathways for the synthesis of secondary metabolites (isoprenoid) and pigments (carotenoid). Keywords: Thermus, Thermophile, Extremophile, Hot spring Introduction Thermus species, Thermus scotoductus SA-01, is also Thermus spp. are Gram-negative, aerobic, non-sporulating, available [6]. T. thermophilus has attracted attention and rod-shaped thermophilic bacteria. Thermus aquaticus as one of the model organisms for structural biology was the first bacterium of the genus Thermus that was studies because protein complexes from extremophiles discovered in several of the hot springs in Yellowstone are easier to crystallize than their mesophilic counter- National Park, United States [1]. -

Annotation Guidelines for Experimental Procedures
Annotation Guidelines for Experimental Procedures Developed By Mohammed Alliheedi Robert Mercer Version 1 April 14th, 2018 1- Introduction and background information What is rhetorical move? A rhetorical move can be defined as a text fragment that conveys a distinct communicative goal, in other words, a sentence that implies an author’s specific purpose to readers. What are the types of rhetorical moves? There are several types of rhetorical moves. However, we are interested in 4 rhetorical moves that are common in the method section of a scientific article that follows the Introduction Methods Results and Discussion (IMRaD) structure. 1- Description of a method: It is concerned with a sentence(s) that describes experimental events (e.g., “Beads with bound proteins were washed six times (for 10 min under rotation at 4°C) with pulldown buffer and proteins harvested in SDS-sample buffer, separated by SDS-PAGE, and analyzed by autoradiography.” (Ester & Uetz, 2008)). 2- Appeal to authority: It is concerned with a sentence(s) that discusses the use of standard methods, protocols, and procedures. There are two types of this move: - A reference to a well-established “standard” method (e.g., the use of a method like “PCR” or “electrophoresis”). - A reference to a method that was previously described in the literature (e.g., “Protein was determined using fluorescamine assay [41].” (Larsen, Frandesn and Treiman, 2001)). 3- Source of materials: It is concerned with a sentence(s) that lists the source of biological materials that are used in the experiment (e.g., “All microalgal strains used in this study are available at the Elizabeth Aidar Microalgae Culture Collection, Department of Marine Biology, Federal Fluminense University, Brazil.” (Larsen, Frandesn and Treiman, 2001)). -

Untargetted Metabolomic Exploration of the Mycobacterium Tuberculosis Stress Response to Cinnamon Essential Oil
biomolecules Article Untargetted Metabolomic Exploration of the Mycobacterium tuberculosis Stress Response to Cinnamon Essential Oil Elwira Sieniawska 1,* , Rafał Sawicki 2 , Joanna Golus 2 and Milen I. Georgiev 3,4 1 Chair and Department of Pharmacognosy, Medical University of Lublin, Chodzki 1, 20-093 Lublin, Poland 2 Chair and Department of Biochemistry and Biotechnology, Medical University of Lublin, Chodzki 1, 20-093 Lublin, Poland; [email protected] (R.S.); [email protected] (J.G.) 3 Group of Plant Cell Biotechnology and Metabolomics, The Stephan Angeloff Institute of Microbiology, Bulgarian Academy of Sciences, 139 Ruski Blvd., 4000 Plovdiv, Bulgaria; [email protected] 4 Center of Plant Systems Biology and Biotechnology, 4000 Plovdiv, Bulgaria * Correspondence: [email protected] Received: 6 January 2020; Accepted: 24 February 2020; Published: 26 February 2020 Abstract: The antimycobacterial activity of cinnamaldehyde has already been proven for laboratory strains and for clinical isolates. What is more, cinnamaldehyde was shown to threaten the mycobacterial plasma membrane integrity and to activate the stress response system. Following promising applications of metabolomics in drug discovery and development we aimed to explore the mycobacteria response to cinnamaldehyde within cinnamon essential oil treatment by untargeted liquid chromatography–mass spectrometry. The use of predictive metabolite pathway analysis and description of produced lipids enabled the evaluation of the stress symptoms shown by bacteria. This study suggests that bacteria exposed to cinnamaldehyde could reorganize their outer membrane as a physical barrier against stress factors. They probably lowered cell wall permeability and inner membrane fluidity, and possibly redirected carbon flow to store energy in triacylglycerols. Being a reactive compound, cinnamaldehyde may also contribute to disturbances in bacteria redox homeostasis and detoxification mechanisms. -

Towards Kinetic Modeling of Global Metabolic Networks with Incomplete Experimental Input on Kinetic Parameters
Towards Kinetic Modeling of Global Metabolic Networks with Incomplete Experimental Input on Kinetic Parameters Ping Ao 1,2 , Lik Wee Lee 1, Mary E. Lidstrom 3,4 , Lan Yin 5, and Xiaomei Zhu 6 1 Department of Mechanical Engineering , University of Washington , Seattle , WA 98195, USA 2 Department of Physics , University of Washington , Seattle , WA 98195, USA 3 Department of Microbiology , University of Washington , Seattle , WA 98195, USA 4 Department of Chemical Engineering , University of Washington , Seattle , WA 98195, USA 5 School of Physics , Peking University , 100871 Beijing , PR China 6 GeneMath , 5525 27 th Ave . N .E., Seattle , WA 98105, USA Abstract: This is the first report on a systematic method for constructing a large scale kinetic metabolic model and its initial application to the modeling of central metabolism of Methylobacterium extorquens AM1, a methylotrophic and environmental important bacterium. Its central metabolic network includes formaldehyde metabolism, serine cycle, citric acid cycle, pentose phosphate pathway, gluconeogensis, PHB synthesis and acetyl-CoA conversion pathway, respiration and energy metabolism. Through a systematic and consistent procedure of finding a set of parameters in the physiological range we overcome an outstanding difficulty in large scale kinetic modeling: the requirement for a massive number of enzymatic reaction parameters. We are able to construct the kinetic model based on general biological considerations and incomplete experimental kinetic parameters. Our method consists of -

Incomplete Denitrification in Thermus Species
UNLV Theses, Dissertations, Professional Papers, and Capstones August 2016 Incomplete Denitrification in Thermus Species Chrisabelle Mefferd University of Nevada, Las Vegas Follow this and additional works at: https://digitalscholarship.unlv.edu/thesesdissertations Part of the Biology Commons, Environmental Sciences Commons, Microbiology Commons, and the Terrestrial and Aquatic Ecology Commons Repository Citation Mefferd, Chrisabelle, "Incomplete Denitrification in Thermus Species" (2016). UNLV Theses, Dissertations, Professional Papers, and Capstones. 2793. http://dx.doi.org/10.34917/9302950 This Thesis is protected by copyright and/or related rights. It has been brought to you by Digital Scholarship@UNLV with permission from the rights-holder(s). You are free to use this Thesis in any way that is permitted by the copyright and related rights legislation that applies to your use. For other uses you need to obtain permission from the rights-holder(s) directly, unless additional rights are indicated by a Creative Commons license in the record and/ or on the work itself. This Thesis has been accepted for inclusion in UNLV Theses, Dissertations, Professional Papers, and Capstones by an authorized administrator of Digital Scholarship@UNLV. For more information, please contact [email protected]. INCOMPLETE DENITRIFICATION IN THERMUS SPECIES By Chrisabelle R. Cempron Bachelor of Sciences - Biology Montclair State University 2013 A thesis submitted in partial fulfillment of the requirements for the Master of Science - Biological Science School of Life Sciences College of Sciences The Graduate College University of Nevada, Las Vegas August 2016 Thesis Approval The Graduate College The University of Nevada, Las Vegas May 25, 2016 This thesis prepared by Chrisabelle R. -

BEING Aquifex Aeolicus: UNTANGLING a HYPERTHERMOPHILE‘S CHECKERED PAST
BEING Aquifex aeolicus: UNTANGLING A HYPERTHERMOPHILE‘S CHECKERED PAST by Robert J.M. Eveleigh Submitted in partial fulfillment of the requirements for the degree of Master of Science at Dalhousie University Halifax, Nova Scotia December 2011 © Copyright by Robert J.M. Eveleigh, 2011 DALHOUSIE UNIVERSITY DEPARTMENT OF COMPUTATIONAL BIOLOGY AND BIOINFORMATICS The undersigned hereby certify that they have read and recommend to the Faculty of Graduate Studies for acceptance a thesis entitled ―BEING Aquifex aeolicus: UNTANGLING A HYPERTHERMOPHILE‘S CHECKERED PAST‖ by Robert J.M. Eveleigh in partial fulfillment of the requirements for the degree of Master of Science. Dated: December 13, 2011 Co-Supervisors: _________________________________ _________________________________ Readers: _________________________________ ii DALHOUSIE UNIVERSITY DATE: December 13, 2011 AUTHOR: Robert J.M. Eveleigh TITLE: BEING Aquifex aeolicus: UNTANGLING A HYPERTHERMOPHILE‘S CHECKERED PAST DEPARTMENT OR SCHOOL: Department of Computational Biology and Bioinformatics DEGREE: MSc CONVOCATION: May YEAR: 2012 Permission is herewith granted to Dalhousie University to circulate and to have copied for non-commercial purposes, at its discretion, the above title upon the request of individuals or institutions. I understand that my thesis will be electronically available to the public. The author reserves other publication rights, and neither the thesis nor extensive extracts from it may be printed or otherwise reproduced without the author‘s written permission. The author attests that permission has been obtained for the use of any copyrighted material appearing in the thesis (other than the brief excerpts requiring only proper acknowledgement in scholarly writing), and that all such use is clearly acknowledged. _______________________________ Signature of Author iii TABLE OF CONTENTS List of Tables .................................................................................................................... -

Effects of Temperature on CRISPR/Cas System Eddie Beckom Augustana College, Rock Island Illinois
Augustana College Augustana Digital Commons Meiothermus ruber Genome Analysis Project Biology 2019 Effects of Temperature on CRISPR/Cas System Eddie Beckom Augustana College, Rock Island Illinois Dr. Lori Scott Augustana College, Rock Island Illinois Follow this and additional works at: https://digitalcommons.augustana.edu/biolmruber Part of the Bioinformatics Commons, Biology Commons, Genomics Commons, and the Molecular Genetics Commons Augustana Digital Commons Citation Beckom, Eddie and Scott, Dr. Lori. "Effects of Temperature on CRISPR/Cas System" (2019). Meiothermus ruber Genome Analysis Project. https://digitalcommons.augustana.edu/biolmruber/45 This Student Paper is brought to you for free and open access by the Biology at Augustana Digital Commons. It has been accepted for inclusion in Meiothermus ruber Genome Analysis Project by an authorized administrator of Augustana Digital Commons. For more information, please contact [email protected]. Eddie Beckom BIO 375 Dr. Lori R. Scott Biology Department, Augustana College 639 38th Street, Rock Island, IL 61201 Temperature Effect on Complexity of CRISPR/Cas Systems What is Meiothermus ruber? Meiothermus ruber is a Gram-negative thermophilic rod-shaped eubacteria . The genus name derives from the Greek words ‘meion’ and ‘thermos’ meaning ‘lesser’ and ‘hot’ to indicate the thermophilic characteristics of Meiothermus ruber. (Nobre et al., 1996; Euzeby, 1997). It lives in thermal environments with an optimal temperature of 60℃. Meiothermus ruber belongs to the bacterial phylum Deinococcus-Thermus. The order Thermales, which is housed within the Thermus group and consists of 6 genera (Vulcanithermus, Oceanithermus, Thermus, Marinithermus, Meiothermus, Rhabdothermus), all containing genera with proteins that are thermostable. (Albuquerque and Costa, 2014). M. ruber is one of eight currently known species in the genus Meiothermus (Euzeby, 1997).