Mouse Slc35c2 Knockout Project (CRISPR/Cas9)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Small Cell Ovarian Carcinoma: Genomic Stability and Responsiveness to Therapeutics
Gamwell et al. Orphanet Journal of Rare Diseases 2013, 8:33 http://www.ojrd.com/content/8/1/33 RESEARCH Open Access Small cell ovarian carcinoma: genomic stability and responsiveness to therapeutics Lisa F Gamwell1,2, Karen Gambaro3, Maria Merziotis2, Colleen Crane2, Suzanna L Arcand4, Valerie Bourada1,2, Christopher Davis2, Jeremy A Squire6, David G Huntsman7,8, Patricia N Tonin3,4,5 and Barbara C Vanderhyden1,2* Abstract Background: The biology of small cell ovarian carcinoma of the hypercalcemic type (SCCOHT), which is a rare and aggressive form of ovarian cancer, is poorly understood. Tumourigenicity, in vitro growth characteristics, genetic and genomic anomalies, and sensitivity to standard and novel chemotherapeutic treatments were investigated in the unique SCCOHT cell line, BIN-67, to provide further insight in the biology of this rare type of ovarian cancer. Method: The tumourigenic potential of BIN-67 cells was determined and the tumours formed in a xenograft model was compared to human SCCOHT. DNA sequencing, spectral karyotyping and high density SNP array analysis was performed. The sensitivity of the BIN-67 cells to standard chemotherapeutic agents and to vesicular stomatitis virus (VSV) and the JX-594 vaccinia virus was tested. Results: BIN-67 cells were capable of forming spheroids in hanging drop cultures. When xenografted into immunodeficient mice, BIN-67 cells developed into tumours that reflected the hypercalcemia and histology of human SCCOHT, notably intense expression of WT-1 and vimentin, and lack of expression of inhibin. Somatic mutations in TP53 and the most common activating mutations in KRAS and BRAF were not found in BIN-67 cells by DNA sequencing. -

Table 2. Significant
Table 2. Significant (Q < 0.05 and |d | > 0.5) transcripts from the meta-analysis Gene Chr Mb Gene Name Affy ProbeSet cDNA_IDs d HAP/LAP d HAP/LAP d d IS Average d Ztest P values Q-value Symbol ID (study #5) 1 2 STS B2m 2 122 beta-2 microglobulin 1452428_a_at AI848245 1.75334941 4 3.2 4 3.2316485 1.07398E-09 5.69E-08 Man2b1 8 84.4 mannosidase 2, alpha B1 1416340_a_at H4049B01 3.75722111 3.87309653 2.1 1.6 2.84852656 5.32443E-07 1.58E-05 1110032A03Rik 9 50.9 RIKEN cDNA 1110032A03 gene 1417211_a_at H4035E05 4 1.66015788 4 1.7 2.82772795 2.94266E-05 0.000527 NA 9 48.5 --- 1456111_at 3.43701477 1.85785922 4 2 2.8237185 9.97969E-08 3.48E-06 Scn4b 9 45.3 Sodium channel, type IV, beta 1434008_at AI844796 3.79536664 1.63774235 3.3 2.3 2.75319499 1.48057E-08 6.21E-07 polypeptide Gadd45gip1 8 84.1 RIKEN cDNA 2310040G17 gene 1417619_at 4 3.38875643 1.4 2 2.69163229 8.84279E-06 0.0001904 BC056474 15 12.1 Mus musculus cDNA clone 1424117_at H3030A06 3.95752801 2.42838452 1.9 2.2 2.62132809 1.3344E-08 5.66E-07 MGC:67360 IMAGE:6823629, complete cds NA 4 153 guanine nucleotide binding protein, 1454696_at -3.46081884 -4 -1.3 -1.6 -2.6026947 8.58458E-05 0.0012617 beta 1 Gnb1 4 153 guanine nucleotide binding protein, 1417432_a_at H3094D02 -3.13334396 -4 -1.6 -1.7 -2.5946297 1.04542E-05 0.0002202 beta 1 Gadd45gip1 8 84.1 RAD23a homolog (S. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Overview of Nucleotide Sugar Transporter Gene Family Functions Across Multiple Species
Review Overview of Nucleotide Sugar Transporter Gene Family Functions Across Multiple Species Ariel Orellana 1,2, Carol Moraga 1, Macarena Araya 1 and Adrian Moreno 1,2 1 - Centro de Biotecnología Vegetal, Universidad Andres Bello, Av. República 217, Santiago, RM 837-0146, Chile 2 - FONDAP Center for Genome Regulation, Santiago, RM,Chile Correspondence to Ariel Orellana: Centro de Biotecnología Vegetal, Universidad Andres Bello, Av. República 217, Santiago, RM 837-0146, Chile. [email protected] http://dx.doi.org/10.1016/j.jmb.2016.05.021 Edited by Thomas J. Smith Abstract Glycoproteins and glycolipids are crucial in a number of cellular processes, such as growth, development, and responses to external cues, among others. Polysaccharides, another class of sugar-containing molecules, also play important structural and signaling roles in the extracellular matrix. The additions of glycans to proteins and lipids, as well as polysaccharide synthesis, are processes that primarily occur in the Golgi apparatus, and the substrates used in this biosynthetic process are nucleotide sugars. These proteins, lipids, and polysaccharides are also modified by the addition of sulfate groups in the Golgi apparatus in a series of reactions where nucleotide sulfate is needed. The required nucleotide sugar substrates are mainly synthesized in the cytosol and transported into the Golgi apparatus by nucleotide sugar transporters (NSTs), which can additionally transport nucleotide sulfate. Due to the critical role of NSTs in eukaryotic organisms, any malfunction of these could change glycan and polysaccharide structures, thus affecting function and altering organism physiology. For example, mutations or deletion on NST genes lead to pathological conditions in humans or alter cell walls in plants. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Epistasis-Driven Identification of SLC25A51 As a Regulator of Human
ARTICLE https://doi.org/10.1038/s41467-020-19871-x OPEN Epistasis-driven identification of SLC25A51 as a regulator of human mitochondrial NAD import Enrico Girardi 1, Gennaro Agrimi 2, Ulrich Goldmann 1, Giuseppe Fiume1, Sabrina Lindinger1, Vitaly Sedlyarov1, Ismet Srndic1, Bettina Gürtl1, Benedikt Agerer 1, Felix Kartnig1, Pasquale Scarcia 2, Maria Antonietta Di Noia2, Eva Liñeiro1, Manuele Rebsamen1, Tabea Wiedmer 1, Andreas Bergthaler1, ✉ Luigi Palmieri2,3 & Giulio Superti-Furga 1,4 1234567890():,; About a thousand genes in the human genome encode for membrane transporters. Among these, several solute carrier proteins (SLCs), representing the largest group of transporters, are still orphan and lack functional characterization. We reasoned that assessing genetic interactions among SLCs may be an efficient way to obtain functional information allowing their deorphanization. Here we describe a network of strong genetic interactions indicating a contribution to mitochondrial respiration and redox metabolism for SLC25A51/MCART1, an uncharacterized member of the SLC25 family of transporters. Through a combination of metabolomics, genomics and genetics approaches, we demonstrate a role for SLC25A51 as enabler of mitochondrial import of NAD, showcasing the potential of genetic interaction- driven functional gene deorphanization. 1 CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences, Vienna, Austria. 2 Laboratory of Biochemistry and Molecular Biology, Department of Biosciences, Biotechnologies and Biopharmaceutics, -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -

Antagonistic Epistasis of Hnf4α and Foxo1 Networks Through Enhancer Interactions
bioRxiv preprint doi: https://doi.org/10.1101/2020.07.04.187864; this version posted July 5, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Kuo et al, Hnf4a and FoxO1 in b-cell function Antagonistic epistasis of Hnf4α and FoxO1 networks through enhancer interactions in β-cell function Taiyi Kuo1,3,*, Wen Du1, Yasutaka Miyachi1, Prasanna K. Dadi2, David A. Jacobson2, Domenico Accili1 1Department of Medicine and Berrie Diabetes Center, Columbia University College of Physicians and Surgeons, New York, NY 2Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, TN 3Lead Contact *Correspondence: Taiyi Kuo, Department of Medicine and Berrie Diabetes Center, Columbia University College of Physicians and Surgeons, 1150 St. Nicholas Avenue, Room 237, New York, New York 10032, USA. Phone: 1-212-851-5333. Email: [email protected]. 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.07.04.187864; this version posted July 5, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Kuo et al, Hnf4a and FoxO1 in b-cell function Abstract Genetic and acquired abnormalities contribute to pancreatic β-cell failure in diabetes. Transcription factors Hnf4α (MODY1) and FoxO1 are respective examples of these two components, and are known to act through β-cell-specific enhancers. -

RNA-Seq Reveals Conservation of Function Among the Yolk Sacs Of
RNA-seq reveals conservation of function among the PNAS PLUS yolk sacs of human, mouse, and chicken Tereza Cindrova-Daviesa, Eric Jauniauxb, Michael G. Elliota,c, Sungsam Gongd,e, Graham J. Burtona,1, and D. Stephen Charnock-Jonesa,d,e,1,2 aCentre for Trophoblast Research, Department of Physiology, Development and Neuroscience, University of Cambridge, Cambridge, CB2 3EG, United Kingdom; bElizabeth Garret Anderson Institute for Women’s Health, Faculty of Population Health Sciences, University College London, London, WC1E 6BT, United Kingdom; cSt. John’s College, University of Cambridge, Cambridge, CB2 1TP, United Kingdom; dDepartment of Obstetrics and Gynaecology, University of Cambridge, Cambridge, CB2 0SW, United Kingdom; and eNational Institute for Health Research, Cambridge Comprehensive Biomedical Research Centre, Cambridge, CB2 0QQ, United Kingdom Edited by R. Michael Roberts, University of Missouri-Columbia, Columbia, MO, and approved May 5, 2017 (received for review February 14, 2017) The yolk sac is phylogenetically the oldest of the extraembryonic yolk sac plays a critical role during organogenesis (3–5, 8–10), membranes. The human embryo retains a yolk sac, which goes there are limited data to support this claim. Obtaining experi- through primary and secondary phases of development, but its mental data for the human is impossible for ethical reasons, and importance is controversial. Although it is known to synthesize thus we adopted an alternative strategy. Here, we report RNA proteins, its transport functions are widely considered vestigial. sequencing (RNA-seq) data derived from human and murine yolk Here, we report RNA-sequencing (RNA-seq) data for the human sacs and compare them with published data from the yolk sac of and murine yolk sacs and compare those data with data for the the chicken. -
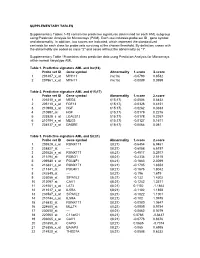
Supplementary Tables 1-18 Contain the Predictive Signatures Determined for Each AML Subgroup Using Prediction Analysis for Microarrays (PAM)
SUPPLEMENTARY TABLES Supplementary Tables 1-18 contain the predictive signatures determined for each AML subgroup using Prediction Analysis for Microarrays (PAM). Each row indicates probe set ID , gene symbol and abnormality. In addition, two scores are indicated, which represent the standardized centroids for each class for probe sets surviving at the chosen threshold. By definition, cases with the abnormality are coded as class "2" and cases without the abnormality as "1". Supplementary Table 19 contains class prediction data using Prediction Analysis for Microarrays within normal karyotype AML. Table 1. Predictive signature AML and inv(16) Probe set ID Gene symbol Abnormality 1.score 2.score 1 201497_x_at MYH11 inv(16) -0.0793 0.8532 2 207961_x_at MYH11 inv(16) -0.0089 0.0959 Table 2. Predictive signature AML and t(15;17) Probe set ID Gene symbol Abnormality 1.score 2.score 1 226210_s_at MEG3 t(15;17) -0.0506 0.6432 2 205110_s_at FGF13 t(15;17) -0.0326 0.4151 3 210998_s_at HGF t(15;17) -0.0262 0.3333 4 210997_at HGF t(15;17) -0.0179 0.2276 5 223828_s_at LGALS12 t(15;17) -0.0178 0.2267 6 210794_s_at MEG3 t(15;17) -0.0127 0.1611 7 204537_s_at GABRE t(15;17) -0.0064 0.081 Table 3. Predictive signature AML and t(8;21) Probe set ID Gene symbol Abnormality 1.score 2.score 1 205529_s_at RUNX1T1 t(8;21) -0.6454 6.9461 2 228827_at --- t(8;21) -0.6058 6.5197 3 205528_s_at RUNX1T1 t(8;21) -0.4917 5.2917 4 213194_at ROBO1 t(8;21) -0.2334 2.5115 5 206940_s_at POU4F1 t(8;21) -0.1883 2.0269 6 216831_s_at RUNX1T1 t(8;21) -0.1705 1.8353 7 211341_at -

Table S5-Correlation of Cytogenetic Changes with Gene Expression
Table S5-Correlation of cytogenetic changes with gene expression in MCF10CA1a FeatureNumCytoband GeneName Description logFC Fold change 28 4087 2p21 TACSTD1 Homo sapiens tumor-associated calcium signal transducer 1 (TACSTD1), mRNA-7.04966 [NM_002354]0.007548158 37 38952 20p12.2 JAG1 Homo sapiens jagged 1 (Alagille syndrome) (JAG1), mRNA [NM_000214] -6.44293 0.011494322 67 2569 20q13.33 COL9A3 Homo sapiens collagen, type IX, alpha 3 (COL9A3), mRNA [NM_001853] -6.04499 0.015145263 72 44151 2p23.3 KRTCAP3 Homo sapiens clone DNA129535 MRV222 (UNQ3066) mRNA, complete cds. [AY358993]-6.03013 0.015302019 105 40016 8q12.1 CA8 Homo sapiens carbonic anhydrase VIII (CA8), mRNA [NM_004056] 5.68101 51.30442111 129 34872 20q13.32 SYCP2 Homo sapiens synaptonemal complex protein 2 (SYCP2), mRNA [NM_014258]-5.21222 0.026975242 140 27988 2p11.2 THC2314643 Unknown -4.99538 0.031350145 149 12276 2p23.3 KRTCAP3 Homo sapiens keratinocyte associated protein 3 (KRTCAP3), mRNA [NM_173853]-4.97604 0.031773429 154 24734 2p11.2 THC2343678 Q6E5T4 (Q6E5T4) Claudin 2, partial (5%) [THC2343678] 5.08397 33.91774866 170 24047 20q13.33 TNFRSF6B Homo sapiens tumor necrosis factor receptor superfamily, member 6b, decoy (TNFRSF6B),-5.09816 0.029194423 transcript variant M68C, mRNA [NM_032945] 177 39605 8p11.21 PLAT Homo sapiens plasminogen activator, tissue (PLAT), transcript variant 1, mRNA-4.88156 [NM_000930]0.033923737 186 31003 20p11.23 OVOL2 Homo sapiens ovo-like 2 (Drosophila) (OVOL2), mRNA [NM_021220] -4.69868 0.038508469 200 39605 8p11.21 PLAT Homo sapiens plasminogen activator, tissue (PLAT), transcript variant 1, mRNA-4.78576 [NM_000930]0.036252773 209 9317 2p14 ARHGAP25 Homo sapiens Rho GTPase activating protein 25 (ARHGAP25), transcript variant-4.62265 1, mRNA0.040592167 [NM_001007231] 211 39605 8p11.21 PLAT Homo sapiens plasminogen activator, tissue (PLAT), transcript variant 1, mRNA -4.7152[NM_000930]0.038070134 212 16979 2p13.1 MGC10955 Homo sapiens hypothetical protein MGC10955, mRNA (cDNA clone MGC:10955-4.70762 IMAGE:3632495),0.038270716 complete cds. -

Autocrine IFN Signaling Inducing Profibrotic Fibroblast Responses By
Downloaded from http://www.jimmunol.org/ by guest on September 23, 2021 Inducing is online at: average * The Journal of Immunology , 11 of which you can access for free at: 2013; 191:2956-2966; Prepublished online 16 from submission to initial decision 4 weeks from acceptance to publication August 2013; doi: 10.4049/jimmunol.1300376 http://www.jimmunol.org/content/191/6/2956 A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Autocrine IFN Signaling Feng Fang, Kohtaro Ooka, Xiaoyong Sun, Ruchi Shah, Swati Bhattacharyya, Jun Wei and John Varga J Immunol cites 49 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2013/08/20/jimmunol.130037 6.DC1 This article http://www.jimmunol.org/content/191/6/2956.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 23, 2021. The Journal of Immunology A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Inducing Autocrine IFN Signaling Feng Fang,* Kohtaro Ooka,* Xiaoyong Sun,† Ruchi Shah,* Swati Bhattacharyya,* Jun Wei,* and John Varga* Activation of TLR3 by exogenous microbial ligands or endogenous injury-associated ligands leads to production of type I IFN.