FASTKD2 Is an RNA-Binding Protein Required for Mitochondrial RNA Processing and Translation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -
H Supplemental Figure 1 I D E B a C
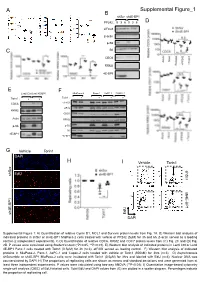
A Supplemental Figure_1 B shScr sh4E-BP1 PP242 : 0 3 6 0 3 6 D eIF4G1 β-actin p-S6 C S6 CDC6 RRM2 4E-BP1 E F Lenti Ctrl Lenti 4E-BP1 MiaPaca-2 Panc-1 AsPC-1 Capan-2 Torin1 : Torin1 : + + + + + + CDC6 eIF4G1 eIF3a RRM2 CDC6 Actin RRM2 p-S6 p-S6 S6 4E-BP1 4E-BP1 G Vehicle Torin1 DAPI H I Vehicle Torin1 62.20% 45.30% shScr sh4E-BP1 EdU shScr 59.58% 49.47% EdU sh4E-BP1 DAPI Supplemental Figure 1: A) Quantification of relative Cyclin D1, MCL1 and Survivin protein levels from Fig. 1A. B) Western blot analysis of indicated proteins in shScr or sh4E-BP1 MiaPaca-2 cells treated with vehicle or PP242 (5µM) for 3h and 6h. β-actin served as a loading control (2 independent experiments). C-D) Quantification of relative CDC6, RRM2 and CDC7 protein levels from (C) Fig. 2C and (D) Fig. 2D. P values were calculated using Student’s t-test (*P<0.05, **P<0.01). E) Western blot analysis of indicated proteins in Lenti Ctrl or Lenti 4E-BP1 Panc-1 cells treated with Torin1 (0.5µM) for 3h (n=3). eIF4GI served as loading control. F) Western blot analysis of indicated proteins in MiaPaca-2, Panc-1, AsPC-1 and Capan-2 cells treated with vehicle or Torin1 (500nM) for 3hrs (n=3). G) Asynchronous shScramble or sh4E-BP1 MiaPaca-2 cells were incubated with Torin1 (0.5μM) for 3hrs and labeled with EdU (n=3). Nuclear DNA was counterstained by DAPI. H) The proportions of replicating cells are shown as means and standard deviations and were generated from at least three independent experiments. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Characterization and Proteomic-Transcriptomic Investigation of Monocarboxylate Transporter 6 Knockout Mice
Supplemental material to this article can be found at: http://molpharm.aspetjournals.org/content/suppl/2019/07/10/mol.119.116731.DC1 1521-0111/96/3/364–376$35.00 https://doi.org/10.1124/mol.119.116731 MOLECULAR PHARMACOLOGY Mol Pharmacol 96:364–376, September 2019 Copyright ª 2019 by The American Society for Pharmacology and Experimental Therapeutics Characterization and Proteomic-Transcriptomic Investigation of Monocarboxylate Transporter 6 Knockout Mice: Evidence of a Potential Role in Glucose and Lipid Metabolism s Robert S. Jones, Chengjian Tu, Ming Zhang, Jun Qu, and Marilyn E. Morris Department of Pharmaceutical Sciences, School of Pharmacy and Pharmaceutical Sciences, University at Buffalo, State University of New York, Buffalo, New York (R.S.J., C.T., J.Q., M.E.M.); and New York State Center of Excellence in Bioinformatics and Life Sciences, Buffalo, New York (C.T., M.Z., J.Q.) Received March 21, 2019; accepted June 27, 2019 Downloaded from ABSTRACT Monocarboxylate transporter 6 [(MCT6), SLC16A5] is an or- Mct62/2 mice demonstrated significant changes in 199 genes phan transporter with no known endogenous substrates or in the liver compared with Mct61/1 mice. In silico biological physiological role. Previous in vitro and in vivo experiments pathway analyses revealed significant changes in proteins and molpharm.aspetjournals.org investigated MCT6 substrate/inhibitor specificity in Xenopus genes involved in glucose and lipid metabolism–associated laevis oocytes; however, these data remain limited. Transcrip- pathways. This study is the first to provide evidence for an tomic changes in the livers of mice undergoing different dieting association of Mct6 in the regulation of glucose and lipid schemes have suggested that Mct6 plays a role in glucose metabolism. -

Gene Ontology Functional Annotations and Pleiotropy
Network based analysis of genetic disease associations Sarah Gilman Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy under the Executive Committee of the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2014 © 2013 Sarah Gilman All Rights Reserved ABSTRACT Network based analysis of genetic disease associations Sarah Gilman Despite extensive efforts and many promising early findings, genome-wide association studies have explained only a small fraction of the genetic factors contributing to common human diseases. There are many theories about where this “missing heritability” might lie, but increasingly the prevailing view is that common variants, the target of GWAS, are not solely responsible for susceptibility to common diseases and a substantial portion of human disease risk will be found among rare variants. Relatively new, such variants have not been subject to purifying selection, and therefore may be particularly pertinent for neuropsychiatric disorders and other diseases with greatly reduced fecundity. Recently, several researchers have made great progress towards uncovering the genetics behind autism and schizophrenia. By sequencing families, they have found hundreds of de novo variants occurring only in affected individuals, both large structural copy number variants and single nucleotide variants. Despite studying large cohorts there has been little recurrence among the genes implicated suggesting that many hundreds of genes may underlie these complex phenotypes. The question -

(Cos) Rnas and of a Conserved Family of Organellar RNA-Binding Proteins, the Heptatricopeptide Repeat Proteins, in the Malaria Parasite Arne Hillebrand1, Joachim M
Published online 8 August 2018 Nucleic Acids Research, 2018, Vol. 46, No. 19 10417–10431 doi: 10.1093/nar/gky710 Identification of clustered organellar short (cos) RNAs and of a conserved family of organellar RNA-binding proteins, the heptatricopeptide repeat proteins, in the malaria parasite Arne Hillebrand1, Joachim M. Matz2, Martin Almendinger1,KatjaMuller¨ 2, Kai Matuschewski2 and Christian Schmitz-Linneweber1,* 1Humboldt University Berlin, Molecular Genetics, Berlin, Germany and 2Humboldt University, Department of Molecular Parasitology, Berlin, Germany Received December 12, 2017; Revised July 20, 2018; Editorial Decision July 23, 2018; Accepted July 24, 2018 ABSTRACT malaria globally, resulting in almost half a million deaths (1). While some antimalarial treatments are available, para- Gene expression in mitochondria of Plasmodium site resistance is a continuing challenge. Plasmodium spp. falciparum is essential for parasite survival. The belong to the family of apicomplexan parasites, most of molecular mechanisms of Plasmodium organellar which contain a non-photosynthetic plastid called the api- gene expression remain poorly understood. This coplast (2). As a remnant of a red algae chloroplast, the api- includes the enigmatic assembly of the mitochon- coplast contains its own DNA, as does the Plasmodium mi- drial ribosome from highly fragmented rRNAs. Here, tochondrion. The Plasmodium nuclear genome has a size we present the identification of clustered organellar of 24 MB and contains >5000 genes; by contrast, the api- short RNA fragments (cosRNAs) that are possible coplast and mitochondrial genomes are small––35 and 6 kb, footprints of RNA-binding proteins (RBPs) in Plas- respectively. modium organelles. In plants, RBPs of the pentatri- In keeping with the descendance of the apicoplast from cyanobacteria, the apicoplast genome is organized similarly copeptide repeat (PPR) class produce footprints as a to bacterial chromosomes. -

A SARS-Cov-2 Protein Interaction Map Reveals Targets for Drug Repurposing
Article A SARS-CoV-2 protein interaction map reveals targets for drug repurposing https://doi.org/10.1038/s41586-020-2286-9 A list of authors and affiliations appears at the end of the paper Received: 23 March 2020 Accepted: 22 April 2020 A newly described coronavirus named severe acute respiratory syndrome Published online: 30 April 2020 coronavirus 2 (SARS-CoV-2), which is the causative agent of coronavirus disease 2019 (COVID-19), has infected over 2.3 million people, led to the death of more than Check for updates 160,000 individuals and caused worldwide social and economic disruption1,2. There are no antiviral drugs with proven clinical efcacy for the treatment of COVID-19, nor are there any vaccines that prevent infection with SARS-CoV-2, and eforts to develop drugs and vaccines are hampered by the limited knowledge of the molecular details of how SARS-CoV-2 infects cells. Here we cloned, tagged and expressed 26 of the 29 SARS-CoV-2 proteins in human cells and identifed the human proteins that physically associated with each of the SARS-CoV-2 proteins using afnity-purifcation mass spectrometry, identifying 332 high-confdence protein–protein interactions between SARS-CoV-2 and human proteins. Among these, we identify 66 druggable human proteins or host factors targeted by 69 compounds (of which, 29 drugs are approved by the US Food and Drug Administration, 12 are in clinical trials and 28 are preclinical compounds). We screened a subset of these in multiple viral assays and found two sets of pharmacological agents that displayed antiviral activity: inhibitors of mRNA translation and predicted regulators of the sigma-1 and sigma-2 receptors. -

Molecular Risk Factors of Pulmonary Arterial Hypertension Hamza M
Florida International University FIU Digital Commons FIU Electronic Theses and Dissertations University Graduate School 9-22-2017 Molecular Risk Factors of Pulmonary Arterial Hypertension Hamza M. Assaggaf Florida International University, [email protected] DOI: 10.25148/etd.FIDC004013 Follow this and additional works at: https://digitalcommons.fiu.edu/etd Part of the Environmental Public Health Commons Recommended Citation Assaggaf, Hamza M., "Molecular Risk Factors of Pulmonary Arterial Hypertension" (2017). FIU Electronic Theses and Dissertations. 3554. https://digitalcommons.fiu.edu/etd/3554 This work is brought to you for free and open access by the University Graduate School at FIU Digital Commons. It has been accepted for inclusion in FIU Electronic Theses and Dissertations by an authorized administrator of FIU Digital Commons. For more information, please contact [email protected]. FLORIDA INTERNATIONAL UNIVERSITY Miami, Florida MOLECULAR RISK FACTORS OF PULMONARY ARTERIAL HYPERTENSION A dissertation submitted in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY in PUBLIC HEALTH by Hamza Assaggaf 2017 To: Dean Tomás R. Guilarte Robert Stempel College of Public Health and Social Work This dissertation, written by Hamza Assaggaf, entitled Molecular Risk Factors of Pulmonary Arterial Hypertension, having been approved in respect to style and intellectual content, is referred to you for judgment. We have read the dissertation and recommend that it be approved. __________________________________________ Alok Deoraj __________________________________________ Juan Luizzi __________________________________________ Deodutta Roy __________________________________________ ChangwonYoo, Co-Major Professor __________________________________________ Quentin Felty, Co-Major Professor Date of Defense: September 22, 2017 The dissertation of Hamza Assaggaf is approved. __________________________________________ Dean Tomás R. Guilarte Robert Stempel College of Public Health and Social Work __________________________________________ Andres G. -

Mitochondrial RNA Granules Are Fluid Condensates, Positioned By
bioRxiv preprint doi: https://doi.org/10.1101/747055; this version posted August 27, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Title: Mitochondrial RNA granules are fluid condensates, positioned by membrane dynamics Authors: Timo Rey1†, Sofia Zaganelli2†, Emilie Cuillery1, Jean-Claude Martinou*, Suliana Manley* 5 Affiliations: 1 Laboratory of Experimental Biophysics, École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland. 2 Department of Cell Biology, University of Geneva, Genève, Switzerland. *Corresponding authors. Email: [email protected]; [email protected]; 10 †these authors contributed equally 15 20 1 bioRxiv preprint doi: https://doi.org/10.1101/747055; this version posted August 27, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Mitochondria contain the genetic information and expression machinery to produce proteins essential for cellular respiration. Within the mitochondrial matrix, newly synthesized RNA, RNA processing proteins, and mitoribosome assembly factors are known to form punctate subcompartments referred to as mitochondrial RNA granules (MRGs) 1-3. 5 Despite their proposed role in regulating gene expression, little is known about the structural and dynamic properties of MRGs. We investigated the organization of MRGs using fluorescence super-resolution localization microscopy and correlative electron microscopy techniques, obtaining ultrastructural details of their internal architecture. -

FASTKD3 Rabbit Polyclonal Antibody – TA349969 | Origene
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TA349969 FASTKD3 Rabbit Polyclonal Antibody Product data: Product Type: Primary Antibodies Applications: WB Recommended Dilution: ELISA: 1000-2000, WB: 200-1000 Reactivity: Human Host: Rabbit Isotype: IgG Clonality: Polyclonal Immunogen: Fusion protein of human FASTKD3 Formulation: pH7.4 PBS, 0.05% NaN3, 40% Glyceroln Concentration: lot specific Purification: Antigen affinity purification Conjugation: Unconjugated Storage: Store at -20°C as received. Stability: Stable for 12 months from date of receipt. Predicted Protein Size: 76 kDa Gene Name: FAST kinase domains 3 Database Link: NP_076996 Entrez Gene 79072 Human Q14CZ7 Background: This gene encodes a member of a small family of Fas-activated serine/threonine kinase domain (FASTKD) containing proteins that share an amino terminal mitochondrial targeting domain and multiple carboxy terminal FAST domains as well as a putative RNA-binding RAP domain. The members of this family are ubiquitously expressed and are generally most abundant in mitochondria-enriched tissues such as heart, skeletal muscle and brown-adipose tissue. Some members of this protein family may play a role in apoptosis. The protein encoded by this gene interacts with components of the mitochondrial respiratory and translation networks. A pseudogene of this gene is also present on chromosome 5. Alternative splicing results -

Induction of Therapeutic Tissue Tolerance Foxp3 Expression Is
Downloaded from http://www.jimmunol.org/ by guest on October 2, 2021 is online at: average * The Journal of Immunology , 13 of which you can access for free at: 2012; 189:3947-3956; Prepublished online 17 from submission to initial decision 4 weeks from acceptance to publication September 2012; doi: 10.4049/jimmunol.1200449 http://www.jimmunol.org/content/189/8/3947 Foxp3 Expression Is Required for the Induction of Therapeutic Tissue Tolerance Frederico S. Regateiro, Ye Chen, Adrian R. Kendal, Robert Hilbrands, Elizabeth Adams, Stephen P. Cobbold, Jianbo Ma, Kristian G. Andersen, Alexander G. Betz, Mindy Zhang, Shruti Madhiwalla, Bruce Roberts, Herman Waldmann, Kathleen F. Nolan and Duncan Howie J Immunol cites 35 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription http://www.jimmunol.org/content/suppl/2012/09/17/jimmunol.120044 9.DC1 This article http://www.jimmunol.org/content/189/8/3947.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2012 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of October 2, 2021. -

Mitochondrial Structure and Bioenergetics in Normal and Disease Conditions
International Journal of Molecular Sciences Review Mitochondrial Structure and Bioenergetics in Normal and Disease Conditions Margherita Protasoni 1 and Massimo Zeviani 1,2,* 1 Mitochondrial Biology Unit, The MRC and University of Cambridge, Cambridge CB2 0XY, UK; [email protected] 2 Department of Neurosciences, University of Padova, 35128 Padova, Italy * Correspondence: [email protected] Abstract: Mitochondria are ubiquitous intracellular organelles found in almost all eukaryotes and involved in various aspects of cellular life, with a primary role in energy production. The interest in this organelle has grown stronger with the discovery of their link to various pathologies, including cancer, aging and neurodegenerative diseases. Indeed, dysfunctional mitochondria cannot provide the required energy to tissues with a high-energy demand, such as heart, brain and muscles, leading to a large spectrum of clinical phenotypes. Mitochondrial defects are at the origin of a group of clinically heterogeneous pathologies, called mitochondrial diseases, with an incidence of 1 in 5000 live births. Primary mitochondrial diseases are associated with genetic mutations both in nuclear and mitochondrial DNA (mtDNA), affecting genes involved in every aspect of the organelle function. As a consequence, it is difficult to find a common cause for mitochondrial diseases and, subsequently, to offer a precise clinical definition of the pathology. Moreover, the complexity of this condition makes it challenging to identify possible therapies or drug targets. Keywords: ATP production; biogenesis of the respiratory chain; mitochondrial disease; mi-tochondrial electrochemical gradient; mitochondrial potential; mitochondrial proton pumping; mitochondrial respiratory chain; oxidative phosphorylation; respiratory complex; respiratory supercomplex Citation: Protasoni, M.; Zeviani, M.