Acyl–Acyl Carrier Protein Desaturases and Plantbiotic Interactions
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Retention Indices for Frequently Reported Compounds of Plant Essential Oils
Retention Indices for Frequently Reported Compounds of Plant Essential Oils V. I. Babushok,a) P. J. Linstrom, and I. G. Zenkevichb) National Institute of Standards and Technology, Gaithersburg, Maryland 20899, USA (Received 1 August 2011; accepted 27 September 2011; published online 29 November 2011) Gas chromatographic retention indices were evaluated for 505 frequently reported plant essential oil components using a large retention index database. Retention data are presented for three types of commonly used stationary phases: dimethyl silicone (nonpolar), dimethyl sili- cone with 5% phenyl groups (slightly polar), and polyethylene glycol (polar) stationary phases. The evaluations are based on the treatment of multiple measurements with the number of data records ranging from about 5 to 800 per compound. Data analysis was limited to temperature programmed conditions. The data reported include the average and median values of retention index with standard deviations and confidence intervals. VC 2011 by the U.S. Secretary of Commerce on behalf of the United States. All rights reserved. [doi:10.1063/1.3653552] Key words: essential oils; gas chromatography; Kova´ts indices; linear indices; retention indices; identification; flavor; olfaction. CONTENTS 1. Introduction The practical applications of plant essential oils are very 1. Introduction................................ 1 diverse. They are used for the production of food, drugs, per- fumes, aromatherapy, and many other applications.1–4 The 2. Retention Indices ........................... 2 need for identification of essential oil components ranges 3. Retention Data Presentation and Discussion . 2 from product quality control to basic research. The identifi- 4. Summary.................................. 45 cation of unknown compounds remains a complex problem, in spite of great progress made in analytical techniques over 5. -

VITAMIN K1 | C31H46O2 - Pubchem
VITAMIN K1 | C31H46O2 - PubChem https://pubchem.ncbi.nlm.nih.gov/compound/Phylloquinone#secti... NIH U.S. National Library of Medicine National Center for Biotechnology Information OPEN CHEMISTRY Search Compounds ! DATABASE VITAMIN K1 " Cite this Record # $ % & ' ( STRUCTURE VENDORS DRUG INFO PHARMACOLOGY LITERATURE PATENTS BIOACTIVITIES PubChem CID: 5284607 VITAMIN K1; Phytonadione; Phylloquinone; 84-80-0; Phytylmenadione; Chemical Names: Phyllochinon More... Molecular Formula: C31H46O2 Molecular Weight: 450.707 g/mol InChI Key: MBWXNTAXLNYFJB-NKFFZRIASA-N Drug Indication Therapeutic Uses Clinical Trials FDA Orange Book Drug Information: FDA UNII Safety Summary: Laboratory Chemical Safety Summary (LCSS) VITAMIN K1 is a family of phylloquinones that contains a ring of 2-methyl-1,4-naphthoquinone and an isoprenoid side chain. Members of this group of vitamin K 1 have only one double bond on the proximal isoprene unit. Rich sources of vitamin K 1 include green plants, algae, and photosynthetic bacteria. Vitamin K1 has antihemorrhagic and prothrombogenic activity. " from MeSH Vitamin K is a family of fat-soluble compounds with a common chemical structure based on 2-methyl-1, 4-naphthoquinone " Metabolite Description from Human Metabolome Database (HMDB) PUBCHEM ) COMPOUND ) VITAMIN K1 Modify Date: 2018-01-06; Create Date: 2004-09-16 1 di 57 12/01/18, 11:36 VITAMIN K1 | C31H46O2 - PubChem https://pubchem.ncbi.nlm.nih.gov/compound/Phylloquinone#secti... * Contents 1 2D Structure 2 3D Conformer 3 Names and Identifiers + 4 Chemical and Physical Properties 5 Related Records 6 Chemical Vendors 7 Drug and Medication Information 8 Pharmacology and Biochemistry 9 Use and Manufacturing 10 Identification 11 Safety and Hazards 12 Toxicity 13 Literature 14 Patents 15 Biomolecular Interactions and Pathways 16 Biological Test Results 17 Classification 18 Information Sources 2 di 57 12/01/18, 11:36 VITAMIN K1 | C31H46O2 - PubChem https://pubchem.ncbi.nlm.nih.gov/compound/Phylloquinone#secti.. -

Contig Protein Description Symbol Anterior Posterior Ratio
Table S2. List of proteins detected in anterior and posterior intestine pooled samples. Data on protein expression are mean ± SEM of 4 pools fed the experimental diets. The number of the contig in the Sea Bream Database (http://nutrigroup-iats.org/seabreamdb) is indicated. Contig Protein Description Symbol Anterior Posterior Ratio Ant/Pos C2_6629 1,4-alpha-glucan-branching enzyme GBE1 0.88±0.1 0.91±0.03 0.98 C2_4764 116 kDa U5 small nuclear ribonucleoprotein component EFTUD2 0.74±0.09 0.71±0.05 1.03 C2_299 14-3-3 protein beta/alpha-1 YWHAB 1.45±0.23 2.18±0.09 0.67 C2_268 14-3-3 protein epsilon YWHAE 1.28±0.2 2.01±0.13 0.63 C2_2474 14-3-3 protein gamma-1 YWHAG 1.8±0.41 2.72±0.09 0.66 C2_1017 14-3-3 protein zeta YWHAZ 1.33±0.14 4.41±0.38 0.30 C2_34474 14-3-3-like protein 2 YWHAQ 1.3±0.11 1.85±0.13 0.70 C2_4902 17-beta-hydroxysteroid dehydrogenase 14 HSD17B14 0.93±0.05 2.33±0.09 0.40 C2_3100 1-acylglycerol-3-phosphate O-acyltransferase ABHD5 ABHD5 0.85±0.07 0.78±0.13 1.10 C2_15440 1-phosphatidylinositol phosphodiesterase PLCD1 0.65±0.12 0.4±0.06 1.65 C2_12986 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta-1 PLCD1 0.76±0.08 1.15±0.16 0.66 C2_4412 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma-2 PLCG2 1.13±0.08 2.08±0.27 0.54 C2_3170 2,4-dienoyl-CoA reductase, mitochondrial DECR1 1.16±0.1 0.83±0.03 1.39 C2_1520 26S protease regulatory subunit 10B PSMC6 1.37±0.21 1.43±0.04 0.96 C2_4264 26S protease regulatory subunit 4 PSMC1 1.2±0.2 1.78±0.08 0.68 C2_1666 26S protease regulatory subunit 6A PSMC3 1.44±0.24 1.61±0.08 -

(Coleoptera) of Peru Miguel A
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Center for Systematic Entomology, Gainesville, Insecta Mundi Florida 2-29-2012 Preliminary checklist of the Cerambycidae, Disteniidae, and Vesperidae (Coleoptera) of Peru Miguel A. Monné Universidade Federal do Rio de Janeiro, [email protected] Eugenio H. Nearns University of New Mexico, [email protected] Sarah C. Carbonel Carril Universidad Nacional Mayor de San Marcos, Peru, [email protected] Ian P. Swift California State Collection of Arthropods, [email protected] Marcela L. Monné Universidade Federal do Rio de Janeiro, [email protected] Follow this and additional works at: http://digitalcommons.unl.edu/insectamundi Part of the Entomology Commons Monné, Miguel A.; Nearns, Eugenio H.; Carbonel Carril, Sarah C.; Swift, Ian P.; and Monné, Marcela L., "Preliminary checklist of the Cerambycidae, Disteniidae, and Vesperidae (Coleoptera) of Peru" (2012). Insecta Mundi. Paper 717. http://digitalcommons.unl.edu/insectamundi/717 This Article is brought to you for free and open access by the Center for Systematic Entomology, Gainesville, Florida at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Insecta Mundi by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. INSECTA MUNDI A Journal of World Insect Systematics 0213 Preliminary checklist of the Cerambycidae, Disteniidae, and Vesperidae (Coleoptera) of Peru Miguel A. Monné Museu Nacional Universidade Federal do Rio de Janeiro Quinta da Boa Vista São Cristóvão, 20940-040 Rio de Janeiro, RJ, Brazil Eugenio H. Nearns Department of Biology Museum of Southwestern Biology University of New Mexico Albuquerque, NM 87131-0001, USA Sarah C. Carbonel Carril Departamento de Entomología Museo de Historia Natural Universidad Nacional Mayor de San Marcos Avenida Arenales 1256, Lima, Peru Ian P. -

Fatty Acid Metabolism and Desaturation in the Pathogenesis of Leukemic Stem Cells in Acute Myeloid Leukemia
Fatty acid metabolism and desaturation in the pathogenesis of leukemic stem cells in Acute Myeloid Leukemia Rachel Culp-Hill1,2, Courtney Jones3, Brett Stevens3, Daniel Pollyea3, Shanshan Pei3, Craig Jordan3, Angelo D’Alessandro1,2 1Graduate Program in Structural Biology and Biochemistry, 2Department of Biochemistry & Molecular Genetics, 3Blood Cancer and BMT Program, Division of Hematology, University of Colorado AMC, Aurora, CO, USA Abstract Ven/Aza targets de novo LSC metabolism… Dysregulation of fatty acid desaturation in relapse p53 controls FA metabolism and desaturation p53 knockout in an AML cell line results in Background: Acute myeloid leukemia (AML) is a cancer of bone marrow- LSCs blasts 510 06 Vehicle Ven+Aza 1.210 7 Saturated Lipids Unsaturated Lipids derived blood cells, where leukemic blasts build up and block function and Newly diagnosed (de novo) ALA ALA increased unsaturated fatty acids, including 1.0 10 7 development of myeloid progenitors. Conventional therapy eliminates bulk LSCs uptake amino acids 410 06 ARG ARG Diagnosis docosapentaenoic acid (22:5) and tumor cells but leukemic stem cells (LSCs) survive, leading to disease ASP more quickly than leukemic 6 ASP 8.010 Relapse docosahexaenoic acid (22:6). Loss of p53 progression and relapse. LSCs uniquely rely on oxidative phosphorylation CYS blasts, and when amino GLU 310 06 6 increases unsaturated fatty acids, similarly (OXPHOS), metabolically driven by amino acid and fatty acid metabolism. C-C GLN 6.010 acids are removed, 06 GLU HIS 210 to the relapsed AML phenotype. 4.010 6 Aim: We have successfully targeted amino acid metabolism in LSCs, but the GLN OXPHOS is reduced. -
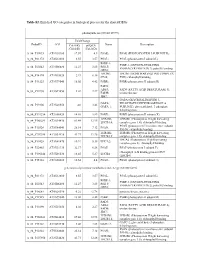
Table S2. Enriched GO Categories in Biological Process for the Shared Degs
Table S2. Enriched GO categories in biological process for the shared DEGs photosynthesis (GO ID:15979) Fold Change ProbeID AGI Col-0(R) pifQ(D) Name Description /Col-0(D) /Col-0(D) A_84_P19035 AT1G30380 17.07 4.9 PSAK; PSAK (PHOTOSYSTEM I SUBUNIT K) A_84_P21372 AT4G12800 8.55 3.57 PSAL; PSAL (photosystem I subunit L) PSBP-1; PSBP-1 (OXYGEN-EVOLVING A_84_P20343 AT1G06680 12.27 3.85 PSII-P; ENHANCER PROTEIN 2); poly(U) binding OEE2; LHCB6; LHCB6 (LIGHT HARVESTING COMPLEX A_84_P14174 AT1G15820 23.9 6.16 CP24; PSII); chlorophyll binding A_84_P11525 AT1G79040 16.02 4.42 PSBR; PSBR (photosystem II subunit R) FAD5; ADS3; FAD5 (FATTY ACID DESATURASE 5); A_84_P19290 AT3G15850 4.02 2.27 FADB; oxidoreductase JB67; GAPA (GLYCERALDEHYDE 3- GAPA; PHOSPHATE DEHYDROGENASE A A_84_P19306 AT3G26650 4.6 3.43 GAPA-1; SUBUNIT); glyceraldehyde-3-phosphate dehydrogenase A_84_P193234 AT2G06520 14.01 3.89 PSBX; PSBX (photosystem II subunit X) LHB1B1; LHB1B1 (Photosystem II light harvesting A_84_P160283 AT2G34430 89.44 32.95 LHCB1.4; complex gene 1.4); chlorophyll binding PSAN (photosystem I reaction center subunit A_84_P10324 AT5G64040 26.14 7.12 PSAN; PSI-N); calmodulin binding LHB1B2; LHB1B2 (Photosystem II light harvesting A_84_P207958 AT2G34420 41.71 12.26 LHCB1.5; complex gene 1.5); chlorophyll binding LHCA2 (Photosystem I light harvesting A_84_P19428 AT3G61470 10.91 5.36 LHCA2; complex gene 2); chlorophyll binding A_84_P22465 AT1G31330 32.37 6.58 PSAF; PSAF (photosystem I subunit F) chlorophyll A-B binding protein CP29 A_84_P190244 AT5G01530 16.45 5.27 LHCB4 -

Fatty Acid Biosynthesis
BI/CH 422/622 ANABOLISM OUTLINE: Photosynthesis Carbon Assimilation – Calvin Cycle Carbohydrate Biosynthesis in Animals Gluconeogenesis Glycogen Synthesis Pentose-Phosphate Pathway Regulation of Carbohydrate Metabolism Anaplerotic reactions Biosynthesis of Fatty Acids and Lipids Fatty Acids contrasts Diversification of fatty acids location & transport Eicosanoids Synthesis Prostaglandins and Thromboxane acetyl-CoA carboxylase Triacylglycerides fatty acid synthase ACP priming Membrane lipids 4 steps Glycerophospholipids Control of fatty acid metabolism Sphingolipids Isoprene lipids: Cholesterol ANABOLISM II: Biosynthesis of Fatty Acids & Lipids 1 ANABOLISM II: Biosynthesis of Fatty Acids & Lipids 1. Biosynthesis of fatty acids 2. Regulation of fatty acid degradation and synthesis 3. Assembly of fatty acids into triacylglycerol and phospholipids 4. Metabolism of isoprenes a. Ketone bodies and Isoprene biosynthesis b. Isoprene polymerization i. Cholesterol ii. Steroids & other molecules iii. Regulation iv. Role of cholesterol in human disease ANABOLISM II: Biosynthesis of Fatty Acids & Lipids Lipid Fat Biosynthesis Catabolism Fatty Acid Fatty Acid Degradation Synthesis Ketone body Isoprene Utilization Biosynthesis 2 Catabolism Fatty Acid Biosynthesis Anabolism • Contrast with Sugars – Lipids have have hydro-carbons not carbo-hydrates – more reduced=more energy – Long-term storage vs short-term storage – Lipids are essential for structure in ALL organisms: membrane phospholipids • Catabolism of fatty acids –produces acetyl-CoA –produces reducing -

Fatty Acid Desaturases: Uncovering Their Involvement in Grapevine Defence Against Downy Mildew
International Journal of Molecular Sciences Article Fatty Acid Desaturases: Uncovering Their Involvement in Grapevine Defence against Downy Mildew Gonçalo Laureano * , Ana Rita Cavaco , Ana Rita Matos and Andreia Figueiredo * Biosystems & Integrative Sciences Institute (BioISI), Faculdade de Ciências, Universidade de Lisboa, Campo Grande, 1749-016 Lisboa, Portugal; [email protected] (A.R.C.); [email protected] (A.R.M.) * Correspondence: [email protected] (G.L.); aafi[email protected] (A.F.) Abstract: Grapevine downy mildew, caused by the biotrophic oomycete Plasmopara viticola, is one of the most severe and devastating diseases in viticulture. Unravelling the grapevine defence mechanisms is crucial to develop sustainable disease control measures. Here we provide new insights concerning fatty acid’s (FA) desaturation, a fundamental process in lipid remodelling and signalling. Previously, we have provided evidence that lipid signalling is essential in the establishment of the incompatible interaction between grapevine and Plasmopara viticola. In the first hours after pathogen challenge, jasmonic acid (JA) accumulation, activation of its biosynthetic pathway and an accumulation of its precursor, the polyunsaturated α-linolenic acid (C18:3), were observed in the leaves of the tolerant genotype, Regent. This work was aimed at a better comprehension of the desaturation processes occurring after inoculation. We characterised, for the first time in Vitis vinifera, the gene family of the FA desaturases and evaluated their involvement in Regent response to Plasmopara viticola. Upon pathogen challenge, an up-regulation of the expression of plastidial FA desaturases genes was observed, resulting in a higher content of polyunsaturated fatty acids (PUFAs) of chloroplast lipids. This study highlights FA desaturases as key players in membrane remodelling Citation: Laureano, G.; Cavaco, A.R.; and signalling in grapevine defence towards biotrophic pathogens. -

Saturated Long-Chain Fatty Acid-Producing Bacteria Contribute
Zhao et al. Microbiome (2018) 6:107 https://doi.org/10.1186/s40168-018-0492-6 RESEARCH Open Access Saturated long-chain fatty acid-producing bacteria contribute to enhanced colonic motility in rats Ling Zhao1†, Yufen Huang2†, Lin Lu1†, Wei Yang1, Tao Huang1, Zesi Lin3, Chengyuan Lin1,4, Hiuyee Kwan1, Hoi Leong Xavier Wong1, Yang Chen5, Silong Sun2, Xuefeng Xie2, Xiaodong Fang2,5, Huanming Yang6, Jian Wang6, Lixin Zhu7* and Zhaoxiang Bian1* Abstract Background: The gut microbiota is closely associated with gastrointestinal (GI) motility disorder, but the mechanism(s) by which bacteria interact with and affect host GI motility remains unclear. In this study, through using metabolomic and metagenomic analyses, an animal model of neonatal maternal separation (NMS) characterized by accelerated colonic motility and gut dysbiosis was used to investigate the mechanism underlying microbiota-driven motility dysfunction. Results: An excess of intracolonic saturated long-chain fatty acids (SLCFAs) was associated with enhanced bowel motility in NMS rats. Heptadecanoic acid (C17:0) and stearic acid (C18:0), as the most abundant odd- and even- numbered carbon SLCFAs in the colon lumen, can promote rat colonic muscle contraction and increase stool frequency. Increase of SLCFAs was positively correlated with elevated abundances of Prevotella, Lactobacillus, and Alistipes. Functional annotation found that the level of bacterial LCFA biosynthesis was highly enriched in NMS group. Essential synthetic genes Fabs were largely identified from the genera Prevotella, Lactobacillus, and Alistipes. Pseudo germ-free (GF) rats receiving fecal microbiota from NMS donors exhibited increased defecation frequency and upregulated bacterial production of intracolonic SLCFAs. Modulation of gut dysbiosis by neomycin effectively attenuated GI motility and reduced bacterial SLCFA generation in the colon lumen of NMS rats. -

Fatty Acids and Stable Isotope Ratios in Shiitake Mushrooms
foods Article Fatty Acids and Stable Isotope Ratios in Shiitake Mushrooms (Lentinula edodes) Indicate the Origin of the Cultivation Substrate Used: A Preliminary Case Study in Korea 1, 1, 2 2 3 Ill-Min Chung y, So-Yeon Kim y, Jae-Gu Han , Won-Sik Kong , Mun Yhung Jung and Seung-Hyun Kim 1,* 1 Department of Crop Science, College of Sanghuh Life Science, Konkuk University, Seoul 05029, Korea; [email protected] (I.-M.C.); [email protected] (S.-Y.K.) 2 National Institute of Horticultural and Herbal Science, Rural Development Administration, Eumseong 27709, Korea; [email protected] (J.-G.H.); [email protected] (W.-S.K.) 3 Department of Food Science and Biotechnology, Graduate School, Woosuk University, Wanju-gun 55338, Korea; [email protected] * Correspondence: [email protected]; Tel.: +82-02-2049-6163; Fax: +82-02-455-1044 These authors contributed equally to this study. y Received: 22 July 2020; Accepted: 28 August 2020; Published: 1 September 2020 Abstract: Shiitake mushroom (Lentinula edodes) is commonly consumed worldwide and is cultivated in many farms in Korea using Chinese substrates owing to a lack of knowledge on how to prepare sawdust-based substrate blocks (bag cultivation). Consequently, issues related to the origin of the Korean or Chinese substrate used in shiitake mushrooms produced using bag cultivation have been reported. Here, we investigated differences in fatty acids (FAs) and stable isotope ratios (SIRs) in shiitake mushrooms cultivated using Korean and Chinese substrates under similar conditions (strain, temperature, humidity, etc.) and depending on the harvesting cycle. The total FA level decreased significantly by 5.49 mg g 1 as the harvesting cycle increased (p < 0.0001); however, no differences · − were found in FAs between shiitake mushrooms cultivated using Korean and Chinese substrates. -

Derived Variables - Nutrients Responses to the 116 Items on the DQES Are Converted to Nutrients by Programs Developed at the CCV
Derived Variables - Nutrients Responses to the 116 items on the DQES are converted to nutrients by programs developed at the CCV. Two standard portion factors used in the calculation of nutrient are included in the data sets. Variable Description PSF Portion Standard Factor APSF a Alcohol Portion Standard Factor Sources of nutrient data Values for most nutrients are based on the Australian food composition data base, NUTTAB95.1 Nutrient values form other sources are listed below. None of these new values have been tested or validated so the CCV recommend that the values be used with caution; the CCV welcome any feedback. o Folate (Folate) and vitamin E (VitE) are derived from British food composition tables.2 o Alpha-Carotene (AlphCarot), Beta-Carotene (Bet_Carot), Beta-Cryptoxanthin (BetaCrypt), Lutein plus Zeaxanthin (LutnZeax) and Lycopene (Lycopene) are derived from the USDA data base3 and are measured in micrograms/day. o Glycemic index (GlycIndex ) and glycemic load (GlycLoad) are derived from an international table.4 Note that the variable Bet_Carot measures total ß-carotene intake, and should not be confused with the variable BetaCarot which estimates ß-carotene equivalents (mcg/day) from NUTTAB and is calculated as the sum of the ß-carotene and half the amounts of - carotene and - and ß-cryptoxanthins present. Although values for the 2 variables are likely to be highly correlated, values may differ to a large extent, having been estimated from different databases, developed at different times in different countries. Also different foods would have been averaged to match values with the DQES items. Hodge at al have described the derivation of values for Glycemic index (GI) and glycemic load (GL) as follows:5 ‘Glycemic index is a method of ranking foods on the basis of the blood glucose response to a given amount of carbohydrate from that food. -

Fatty Acid Desaturases, Polyunsaturated Fatty Acid Regulation, and Biotechnological Advances
Review Fatty Acid Desaturases, Polyunsaturated Fatty Acid Regulation, and Biotechnological Advances Je Min Lee 1,†, Hyungjae Lee 2,†, SeokBeom Kang 3 and Woo Jung Park 4,* Received: 11 October 2015; Accepted: 17 December 2015; Published: 4 January 2016 1 Department of Horticultural Science, Kyungpook National University, Daegu 41566, Korea; [email protected] 2 Department of Food Engineering, Dankook University, Cheonan, Chungnam 31116, Korea; [email protected] 3 Citrus Research Station, National Institute of Horticultural & Herbal Science, RDA, Seogwipo 63607, Korea; [email protected] 4 Department of Marine Food Science and Technology, Gangneung-Wonju National University, Gangneung, Gangwon 25457, Korea * Correspondence: [email protected] or [email protected]; Tel.: +82-33-640-2857; Fax: +82-33-640-2850 † These authors contributed equally to this work. Abstract: Polyunsaturated fatty acids (PUFAs) are considered to be critical nutrients to regulate human health and development, and numerous fatty acid desaturases play key roles in synthesizing PUFAs. Given the lack of delta-12 and -15 desaturases and the low levels of conversion to PUFAs, humans must consume some omega-3 and omega-6 fatty acids in their diet. Many studies on fatty acid desaturases as well as PUFAs have shown that fatty acid desaturase genes are closely related to different human physiological conditions. Since the first front-end desaturases from cyanobacteria were cloned, numerous desaturase genes have been identified and animals and plants have been genetically engineered to produce PUFAs such as eicosapentaenoic acid and docosahexaenoic acid. Recently, a biotechnological approach has been used to develop clinical treatments for human physiological conditions, including cancers and neurogenetic disorders.